Gene
KWMTBOMO06995 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010478
Annotation
PREDICTED:_protein_FAM151A_isoform_X2_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.127
Sequence
CDS
ATGTTTCTTGTCATAATCCTGTTTGTCACAGCTACATTCGTGACCTGTGAAGAAGAAACTATGAAGAACTTAACAACAGTTACTTGGGCGCACGCCGTCAACAACAAAACATATTTGGAAGCTGCCTTAGCAAGTGAAGTTGCAATGCTAGAAGCCGACATTGTTTTGGGACATCTAACAGGCCACGACGGTCCTCCAATACCAATAATGGCTCATCCTCCAGCAACCACGTCAGATCTGTCATTGGCTGAATTTCTGTCGACGGTGGCACAATATAACAACGTTAATAATAAACAGAAAGGAGTTAAGCTTGACTTTAAAAGTATTGAAGTGTTTTCTAAGAGCCAGGAGCTGATCGCTCCTTACAGTAAACCTCAGGTGACTTTCCCATTATGGTTAAATGCCGACATTCTACCTGGTCCAGTAAAAGCGACAACAACCCCCGTTGATCCCATCAAATTCATAGAACTAGGTGCGAATCATCCTCGCGCTGTGCTATGTATGGGCTGGACCACTAGATACGGTGGTAACATCACCGAGGGGGAGTATACTCCTGAACAAATTGGTTCAATGCTGAGATTAGTCAATGAACAGAAAATCAATCAGACTATCACATTCCCAGTGCGCGCAGGTCTTGCCTGCAATAGTCAACCGGTACTTTTGGACCTGCTTCGGGAGACAGCGTCCTTGAATTCCTCAATGACTGTCTGGTCCAGCGAAGGAGACTCCGTGGAGGTCGGCCGTCTGAAGGCTCTCATCCTGACCGTGGGACTCGAAAGGACATACTTGGACGTACCACACGATCTGGCTGCCAAACTGAACCTACCTTCGACTGATAAAGTCAAACATTAA
Protein
MFLVIILFVTATFVTCEEETMKNLTTVTWAHAVNNKTYLEAALASEVAMLEADIVLGHLTGHDGPPIPIMAHPPATTSDLSLAEFLSTVAQYNNVNNKQKGVKLDFKSIEVFSKSQELIAPYSKPQVTFPLWLNADILPGPVKATTTPVDPIKFIELGANHPRAVLCMGWTTRYGGNITEGEYTPEQIGSMLRLVNEQKINQTITFPVRAGLACNSQPVLLDLLRETASLNSSMTVWSSEGDSVEVGRLKALILTVGLERTYLDVPHDLAAKLNLPSTDKVKH
Summary
Similarity
Belongs to the CD36 family.
Belongs to the CRISP family.
Belongs to the CRISP family.
Uniprot
H9JLS6
A0A1E1WCD7
A0A212EQ98
A0A194QPF2
A0A1E1VYP6
A0A3S2LNN1
+ More
A0A194QJD4 A0A2A4K527 A0A2H1V3C7 A0A2W1BCJ6 A0A0L7LNC5 A0A1W4WU94 A0A1W4X541 A0A1W4WU70 A0A1W4X3S2 A0A1W4X4Z1 B4JR24 T1PNI6 T1PEH0 A0A1L8EIL8 A0A1L8EIP2 A0A1L8EIT5 A0A0P8XHP1 A0A1L8EIY7 A0A0J7KZQ0 A0A1L8D9G2 A0A1L8D9G9 A0A1I8MAX4 B3MVG1 J3JWA7 E2BT85 A0A1I8Q762 B4LQK3 B4G867 B4MVY2 Q29N66 A0A1L8D9S5 A0A0T6AUV1 A0A1I8Q766 A0A0R3NS43 A0A0Q9WBH3 A0A0M4E8P0 A0A034WML0 A0A3B0K2L3 A0A026X3M0 A0A034WLP4 Q8MQP2 A0A3B0JCQ8 G7H7Z7 A0A034WN28 A0A1B0CRZ9 A0A1B6DAG5 Q9VLU9 K7J1P3 B4KJ93 A0A0L7RE72 A0A0J9TIA4 A0A1W4W7E5 W8CB12 B4Q677 A0A2A3EIU7 E9IYM2 A0A2M4BWN8 A0A0Q9X5Q7 A0A0R1DS61 A0A0K8WBG1 B4NWB2 A0A0Q9X544 B4HYC5 A0A1W4VWE5 V9IDE0 A0A088AN34 A0A232ET29 A0A139WJR4 E2B0D3 D2A1M8 A0A0Q5WBW8 A0A1A9WDN6 W5J3N5 A0A2M4AUI0 A0A2M4AUP0 B3N6R2 A0A182MNB7 A0A0M8ZW16 A0A0L0CFL9 A0A067RWF0 A0A336MFA4 A0A1Y1N968 A0A1J0M5I8 Q7Q3G4 U5EVK7 A0A182UB38 A0A182L2A3 A0A1S4GWG2 A0A182SEN3 U4UM28 A0A2P8YIR6 A0A336MEX1 U4UDU5 D3TLN5
A0A194QJD4 A0A2A4K527 A0A2H1V3C7 A0A2W1BCJ6 A0A0L7LNC5 A0A1W4WU94 A0A1W4X541 A0A1W4WU70 A0A1W4X3S2 A0A1W4X4Z1 B4JR24 T1PNI6 T1PEH0 A0A1L8EIL8 A0A1L8EIP2 A0A1L8EIT5 A0A0P8XHP1 A0A1L8EIY7 A0A0J7KZQ0 A0A1L8D9G2 A0A1L8D9G9 A0A1I8MAX4 B3MVG1 J3JWA7 E2BT85 A0A1I8Q762 B4LQK3 B4G867 B4MVY2 Q29N66 A0A1L8D9S5 A0A0T6AUV1 A0A1I8Q766 A0A0R3NS43 A0A0Q9WBH3 A0A0M4E8P0 A0A034WML0 A0A3B0K2L3 A0A026X3M0 A0A034WLP4 Q8MQP2 A0A3B0JCQ8 G7H7Z7 A0A034WN28 A0A1B0CRZ9 A0A1B6DAG5 Q9VLU9 K7J1P3 B4KJ93 A0A0L7RE72 A0A0J9TIA4 A0A1W4W7E5 W8CB12 B4Q677 A0A2A3EIU7 E9IYM2 A0A2M4BWN8 A0A0Q9X5Q7 A0A0R1DS61 A0A0K8WBG1 B4NWB2 A0A0Q9X544 B4HYC5 A0A1W4VWE5 V9IDE0 A0A088AN34 A0A232ET29 A0A139WJR4 E2B0D3 D2A1M8 A0A0Q5WBW8 A0A1A9WDN6 W5J3N5 A0A2M4AUI0 A0A2M4AUP0 B3N6R2 A0A182MNB7 A0A0M8ZW16 A0A0L0CFL9 A0A067RWF0 A0A336MFA4 A0A1Y1N968 A0A1J0M5I8 Q7Q3G4 U5EVK7 A0A182UB38 A0A182L2A3 A0A1S4GWG2 A0A182SEN3 U4UM28 A0A2P8YIR6 A0A336MEX1 U4UDU5 D3TLN5
Pubmed
19121390
22118469
26354079
28756777
26227816
17994087
+ More
25315136 22516182 20798317 15632085 25348373 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 22936249 24495485 21282665 18057021 17550304 28648823 18362917 19820115 20920257 23761445 26108605 24845553 28004739 12364791 14747013 17210077 20966253 23537049 29403074 20353571
25315136 22516182 20798317 15632085 25348373 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 22936249 24495485 21282665 18057021 17550304 28648823 18362917 19820115 20920257 23761445 26108605 24845553 28004739 12364791 14747013 17210077 20966253 23537049 29403074 20353571
EMBL
BABH01005162
GDQN01006400
JAT84654.1
AGBW02013290
OWR43647.1
KQ461194
+ More
KPJ06855.1 GDQN01011242 JAT79812.1 RSAL01000046 RVE50584.1 KQ458671 KPJ05504.1 NWSH01000120 PCG79337.1 ODYU01000476 SOQ35331.1 KZ150510 PZC70640.1 JTDY01000553 KOB76696.1 CH916372 EDV99354.1 KA649650 AFP64279.1 KA646520 AFP61149.1 GFDG01000209 JAV18590.1 GFDG01000207 JAV18592.1 GFDG01000208 JAV18591.1 CH902624 KPU74360.1 GFDG01000210 JAV18589.1 LBMM01001726 KMQ95841.1 GFDF01010975 JAV03109.1 GFDF01010976 JAV03108.1 EDV33226.2 BT127525 AEE62487.1 GL450384 EFN81087.1 CH940649 EDW64460.1 CH479180 EDW28547.1 CH963857 EDW75852.1 CH379060 EAL33477.2 GFDF01010977 JAV03107.1 LJIG01022764 KRT78838.1 KRT03876.1 KRF81638.1 CP012523 ALC39340.1 GAKP01003375 JAC55577.1 OUUW01000004 SPP79846.1 KK107013 QOIP01000003 EZA62872.1 RLU25060.1 GAKP01003378 GAKP01003373 JAC55579.1 AE014134 AY128469 BT120201 AAF52583.3 AAM75062.1 ADB91449.1 SPP79845.1 BT132720 AER29910.1 GAKP01003377 JAC55575.1 AJWK01025520 GEDC01014688 JAS22610.1 AAF52582.2 CH933807 EDW12468.1 KQ414613 KOC69026.1 CM002910 KMY88885.1 GAMC01000864 JAC05692.1 CM000361 EDX04153.1 KMY88884.1 KZ288245 PBC31202.1 GL767043 EFZ14328.1 GGFJ01007997 MBW57138.1 KRG03261.1 KRG03262.1 CM000157 KRJ97779.1 GDHF01003940 JAI48374.1 EDW88429.1 KRG03263.1 CH480818 EDW52055.1 JR040311 JR040312 AEY59080.1 AEY59081.1 NNAY01002342 OXU21511.1 KQ971338 KYB28162.1 GL444507 EFN60852.1 EFA02695.2 CH954177 KQS70724.1 ADMH02002133 ETN58451.1 GGFK01011112 MBW44433.1 GGFK01011111 MBW44432.1 EDV59278.1 AXCM01009728 KQ435824 KOX72097.1 JRES01000438 KNC31208.1 KK852424 KDR24219.1 UFQS01001089 UFQT01001089 SSX08945.1 SSX28856.1 GEZM01014351 JAV92137.1 KU663642 APD15636.1 AAAB01008964 EAA12160.3 GANO01003362 JAB56509.1 KB632278 ERL91226.1 PYGN01000566 PSN44146.1 SSX08944.1 SSX28855.1 ERL91227.1 EZ422337 ADD18613.1
KPJ06855.1 GDQN01011242 JAT79812.1 RSAL01000046 RVE50584.1 KQ458671 KPJ05504.1 NWSH01000120 PCG79337.1 ODYU01000476 SOQ35331.1 KZ150510 PZC70640.1 JTDY01000553 KOB76696.1 CH916372 EDV99354.1 KA649650 AFP64279.1 KA646520 AFP61149.1 GFDG01000209 JAV18590.1 GFDG01000207 JAV18592.1 GFDG01000208 JAV18591.1 CH902624 KPU74360.1 GFDG01000210 JAV18589.1 LBMM01001726 KMQ95841.1 GFDF01010975 JAV03109.1 GFDF01010976 JAV03108.1 EDV33226.2 BT127525 AEE62487.1 GL450384 EFN81087.1 CH940649 EDW64460.1 CH479180 EDW28547.1 CH963857 EDW75852.1 CH379060 EAL33477.2 GFDF01010977 JAV03107.1 LJIG01022764 KRT78838.1 KRT03876.1 KRF81638.1 CP012523 ALC39340.1 GAKP01003375 JAC55577.1 OUUW01000004 SPP79846.1 KK107013 QOIP01000003 EZA62872.1 RLU25060.1 GAKP01003378 GAKP01003373 JAC55579.1 AE014134 AY128469 BT120201 AAF52583.3 AAM75062.1 ADB91449.1 SPP79845.1 BT132720 AER29910.1 GAKP01003377 JAC55575.1 AJWK01025520 GEDC01014688 JAS22610.1 AAF52582.2 CH933807 EDW12468.1 KQ414613 KOC69026.1 CM002910 KMY88885.1 GAMC01000864 JAC05692.1 CM000361 EDX04153.1 KMY88884.1 KZ288245 PBC31202.1 GL767043 EFZ14328.1 GGFJ01007997 MBW57138.1 KRG03261.1 KRG03262.1 CM000157 KRJ97779.1 GDHF01003940 JAI48374.1 EDW88429.1 KRG03263.1 CH480818 EDW52055.1 JR040311 JR040312 AEY59080.1 AEY59081.1 NNAY01002342 OXU21511.1 KQ971338 KYB28162.1 GL444507 EFN60852.1 EFA02695.2 CH954177 KQS70724.1 ADMH02002133 ETN58451.1 GGFK01011112 MBW44433.1 GGFK01011111 MBW44432.1 EDV59278.1 AXCM01009728 KQ435824 KOX72097.1 JRES01000438 KNC31208.1 KK852424 KDR24219.1 UFQS01001089 UFQT01001089 SSX08945.1 SSX28856.1 GEZM01014351 JAV92137.1 KU663642 APD15636.1 AAAB01008964 EAA12160.3 GANO01003362 JAB56509.1 KB632278 ERL91226.1 PYGN01000566 PSN44146.1 SSX08944.1 SSX28855.1 ERL91227.1 EZ422337 ADD18613.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000283053
UP000053268
UP000218220
+ More
UP000037510 UP000192223 UP000001070 UP000007801 UP000036403 UP000095301 UP000008237 UP000095300 UP000008792 UP000008744 UP000007798 UP000001819 UP000092553 UP000268350 UP000053097 UP000279307 UP000000803 UP000092461 UP000002358 UP000009192 UP000053825 UP000192221 UP000000304 UP000242457 UP000002282 UP000001292 UP000005203 UP000215335 UP000007266 UP000000311 UP000008711 UP000091820 UP000000673 UP000075883 UP000053105 UP000037069 UP000027135 UP000007062 UP000075902 UP000075882 UP000075901 UP000030742 UP000245037
UP000037510 UP000192223 UP000001070 UP000007801 UP000036403 UP000095301 UP000008237 UP000095300 UP000008792 UP000008744 UP000007798 UP000001819 UP000092553 UP000268350 UP000053097 UP000279307 UP000000803 UP000092461 UP000002358 UP000009192 UP000053825 UP000192221 UP000000304 UP000242457 UP000002282 UP000001292 UP000005203 UP000215335 UP000007266 UP000000311 UP000008711 UP000091820 UP000000673 UP000075883 UP000053105 UP000037069 UP000027135 UP000007062 UP000075902 UP000075882 UP000075901 UP000030742 UP000245037
PRIDE
Interpro
SUPFAM
SSF55797
SSF55797
Gene 3D
ProteinModelPortal
H9JLS6
A0A1E1WCD7
A0A212EQ98
A0A194QPF2
A0A1E1VYP6
A0A3S2LNN1
+ More
A0A194QJD4 A0A2A4K527 A0A2H1V3C7 A0A2W1BCJ6 A0A0L7LNC5 A0A1W4WU94 A0A1W4X541 A0A1W4WU70 A0A1W4X3S2 A0A1W4X4Z1 B4JR24 T1PNI6 T1PEH0 A0A1L8EIL8 A0A1L8EIP2 A0A1L8EIT5 A0A0P8XHP1 A0A1L8EIY7 A0A0J7KZQ0 A0A1L8D9G2 A0A1L8D9G9 A0A1I8MAX4 B3MVG1 J3JWA7 E2BT85 A0A1I8Q762 B4LQK3 B4G867 B4MVY2 Q29N66 A0A1L8D9S5 A0A0T6AUV1 A0A1I8Q766 A0A0R3NS43 A0A0Q9WBH3 A0A0M4E8P0 A0A034WML0 A0A3B0K2L3 A0A026X3M0 A0A034WLP4 Q8MQP2 A0A3B0JCQ8 G7H7Z7 A0A034WN28 A0A1B0CRZ9 A0A1B6DAG5 Q9VLU9 K7J1P3 B4KJ93 A0A0L7RE72 A0A0J9TIA4 A0A1W4W7E5 W8CB12 B4Q677 A0A2A3EIU7 E9IYM2 A0A2M4BWN8 A0A0Q9X5Q7 A0A0R1DS61 A0A0K8WBG1 B4NWB2 A0A0Q9X544 B4HYC5 A0A1W4VWE5 V9IDE0 A0A088AN34 A0A232ET29 A0A139WJR4 E2B0D3 D2A1M8 A0A0Q5WBW8 A0A1A9WDN6 W5J3N5 A0A2M4AUI0 A0A2M4AUP0 B3N6R2 A0A182MNB7 A0A0M8ZW16 A0A0L0CFL9 A0A067RWF0 A0A336MFA4 A0A1Y1N968 A0A1J0M5I8 Q7Q3G4 U5EVK7 A0A182UB38 A0A182L2A3 A0A1S4GWG2 A0A182SEN3 U4UM28 A0A2P8YIR6 A0A336MEX1 U4UDU5 D3TLN5
A0A194QJD4 A0A2A4K527 A0A2H1V3C7 A0A2W1BCJ6 A0A0L7LNC5 A0A1W4WU94 A0A1W4X541 A0A1W4WU70 A0A1W4X3S2 A0A1W4X4Z1 B4JR24 T1PNI6 T1PEH0 A0A1L8EIL8 A0A1L8EIP2 A0A1L8EIT5 A0A0P8XHP1 A0A1L8EIY7 A0A0J7KZQ0 A0A1L8D9G2 A0A1L8D9G9 A0A1I8MAX4 B3MVG1 J3JWA7 E2BT85 A0A1I8Q762 B4LQK3 B4G867 B4MVY2 Q29N66 A0A1L8D9S5 A0A0T6AUV1 A0A1I8Q766 A0A0R3NS43 A0A0Q9WBH3 A0A0M4E8P0 A0A034WML0 A0A3B0K2L3 A0A026X3M0 A0A034WLP4 Q8MQP2 A0A3B0JCQ8 G7H7Z7 A0A034WN28 A0A1B0CRZ9 A0A1B6DAG5 Q9VLU9 K7J1P3 B4KJ93 A0A0L7RE72 A0A0J9TIA4 A0A1W4W7E5 W8CB12 B4Q677 A0A2A3EIU7 E9IYM2 A0A2M4BWN8 A0A0Q9X5Q7 A0A0R1DS61 A0A0K8WBG1 B4NWB2 A0A0Q9X544 B4HYC5 A0A1W4VWE5 V9IDE0 A0A088AN34 A0A232ET29 A0A139WJR4 E2B0D3 D2A1M8 A0A0Q5WBW8 A0A1A9WDN6 W5J3N5 A0A2M4AUI0 A0A2M4AUP0 B3N6R2 A0A182MNB7 A0A0M8ZW16 A0A0L0CFL9 A0A067RWF0 A0A336MFA4 A0A1Y1N968 A0A1J0M5I8 Q7Q3G4 U5EVK7 A0A182UB38 A0A182L2A3 A0A1S4GWG2 A0A182SEN3 U4UM28 A0A2P8YIR6 A0A336MEX1 U4UDU5 D3TLN5
Ontologies
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.986849
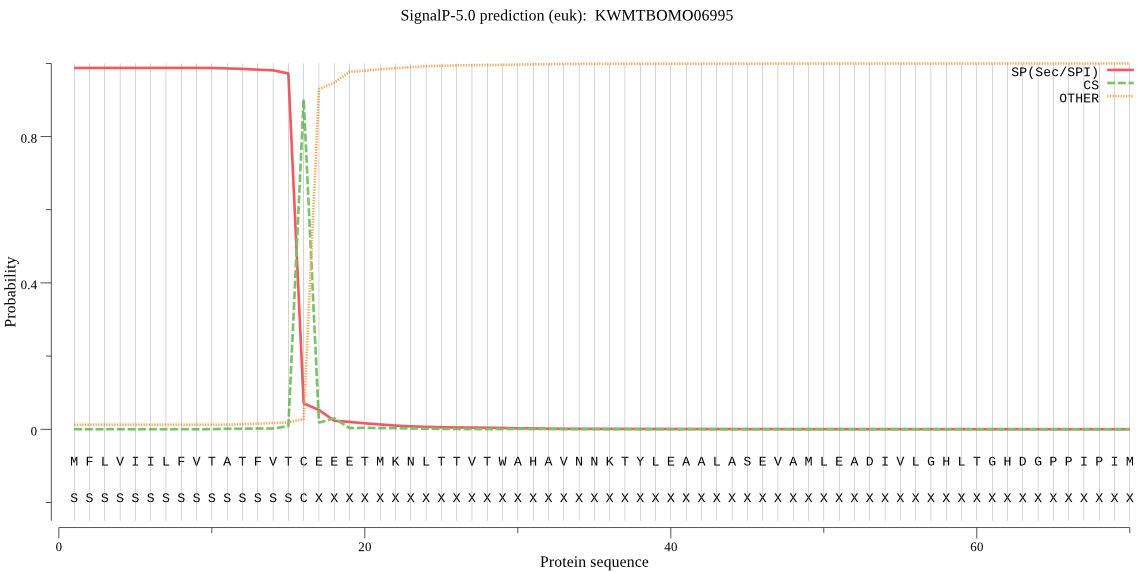
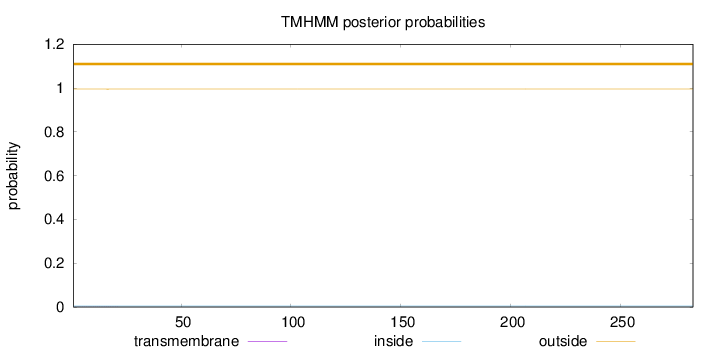
Length:
283
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01323
Exp number, first 60 AAs:
0.01298
Total prob of N-in:
0.00481
outside
1 - 283
Population Genetic Test Statistics
Pi
213.066835
Theta
195.329617
Tajima's D
0.261838
CLR
0.621392
CSRT
0.43962801859907
Interpretation
Uncertain
