Gene
KWMTBOMO06990
Pre Gene Modal
BGIBMGA010501
Annotation
PREDICTED:_uncharacterized_protein_LOC101737072_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.32
Sequence
CDS
ATGATGCTCCTCCTGCTCATCGCGTGTGTCGCAGTTACAACTGCCGAAAATGCCACTAAAGATTCAACCGAACCACCTTCGAACACCACTAAAGAGATCTTGAAAGCTAGTGAAACAGCTGATGCATCAAAAGTTGAAATAGTCAAGCAAATAAGGAGACTTAACGACGACGGGTCTTATACTATTGGATATGAGGCTGACGACGGTACGTTTAAGATTGAGAGTAGGGATGTGCGCGGTAATGTGAAGGGAACTTTTGGATATATCGACAAGGATGGTGAAATCAAACGGGTGACGTACTCGTCTTCTGGGGAAAGCACTCCGATCCCGGTGACAACAACCACAACGACCACGCAAAGCTCCCCAACGATGATAGTGAGAGTGAATAAAACTGTGTCGTCGACTACAAGGAGACCACTAGCCACTGTGGTGTATCCGACTCGGAGTAGCTTCACGACTAGAGGTACCGTAATTCAATCTATACCGAGAAGGAGAGGCTCCAGTACGAGACCTCCAAACACAACCGAATCAACGACTGAGCAACAGAATCTCGAGGATACAACCAGCAGCATTTTGAATTTGTACAAACCGAAAGTCGAGGAGAACATTAGAACGAGAACACAAATATTAAAACCCATGTCAGATATACTTAAAGACGACCTGGTGACAAGGCAGACTACGACCAAATTGCCGCAAACCGTGAAGCCCGTTTATGAGCACGTAACCGAAAAGGCACCGGAGAGTGTGCTGAAGATGAACACATTTCGAAGAGAGTTACCAAGTACGACTACTAACCCTCATATGATCAACCTTCAGCAGTCAGCTGGTGATGATTCCACTGACGAATACGGAAGTCAGCTGTCACTTGGAACTATACGTCCATTGTTTACGACGACCACTCCACGACCTCGATTGGTTCCTATTCAATCAATAGTGGCAGCAAGGCAGAGAATACAGCAACGCTACCAGGATGCACCTCTGGATGACCAAGAAACAACGATAGAAGCAAGCACAGGCAGAGTCTTTGATAGTCAAGGTGTCGTGACTTCTAATCCGCTTCCAGTTGTTCATATACCAGCACAGAGAGAACCTGATGACCGCGCGTATCAGCAAACAGTATATCGAAGACCGCCAGCAGTTCAGTTCCGCACTCGGGAATATTTGCGAGATAATCCCGGAGCTGCCGTACCTATAGGTAACCAGCGGCCCTTTCTCCACTACGAATACGAGAAAGTACTGGAGCCTCAATACGATAAGGAGCCAGTCCAGACTCTTCAGAAACCAGAACCGACCGCAGCGTCATATGACACTAGACCTGTGACGCGAATTGTTCCTATACCGGTTGACGAAAGAGGCGTTCCAATACCGGGGTATCAGGCAAGGTTCGCTAACGCGTATAGACCGCAGCCAGCTCTCTTCCAGCCTCGGTATGAGTCCGTAGATGATATGCACAGTATATCAGCACCAGTTAGTACACGGGACTTAAGACGTTTACTGTACATTCTTATTCTGAGGCAGAACCGATTGCAGGCGCTTGTGGACCAAATGATGCCAGGATCTGCGTACCCAATGCCGATCTACAGAGACAGTTATAGATACCAAAATCGTTACGAAGATGATGGATACGATTATAGATACGATCCAAAATATAGGAACGATTACATGCAGCCCATGTATGAAAATCAATATGAGAACCATCGGTACTTGCCTAGAAGGAGGATGTATCCGCGTCCTTATGACACTGCAGGTTCAGCCTCGAACCACATTGAGGAAGTTCCGGAATTCCTGCCACCAGAAGTAAGAGAGGCATTGCTTCTGAAGATGTTGATTTTAGCTATTAGTCCGGATTTCGCGCCGACTCCTGTGCCGCCCACGGAATTGACCACTGCTGCTCCAATGAGAAAACAAGTAAGAAACGTTCAGATACTTGGAGAAGAGGGCCCTATGACTGATAAACAGAGACATTGA
Protein
MMLLLLIACVAVTTAENATKDSTEPPSNTTKEILKASETADASKVEIVKQIRRLNDDGSYTIGYEADDGTFKIESRDVRGNVKGTFGYIDKDGEIKRVTYSSSGESTPIPVTTTTTTTQSSPTMIVRVNKTVSSTTRRPLATVVYPTRSSFTTRGTVIQSIPRRRGSSTRPPNTTESTTEQQNLEDTTSSILNLYKPKVEENIRTRTQILKPMSDILKDDLVTRQTTTKLPQTVKPVYEHVTEKAPESVLKMNTFRRELPSTTTNPHMINLQQSAGDDSTDEYGSQLSLGTIRPLFTTTTPRPRLVPIQSIVAARQRIQQRYQDAPLDDQETTIEASTGRVFDSQGVVTSNPLPVVHIPAQREPDDRAYQQTVYRRPPAVQFRTREYLRDNPGAAVPIGNQRPFLHYEYEKVLEPQYDKEPVQTLQKPEPTAASYDTRPVTRIVPIPVDERGVPIPGYQARFANAYRPQPALFQPRYESVDDMHSISAPVSTRDLRRLLYILILRQNRLQALVDQMMPGSAYPMPIYRDSYRYQNRYEDDGYDYRYDPKYRNDYMQPMYENQYENHRYLPRRRMYPRPYDTAGSASNHIEEVPEFLPPEVREALLLKMLILAISPDFAPTPVPPTELTTAAPMRKQVRNVQILGEEGPMTDKQRH
Summary
Uniprot
EMBL
Proteomes
Pfam
PF00379 Chitin_bind_4
Interpro
IPR000618
Insect_cuticle
ProteinModelPortal
Ontologies
GO
Topology
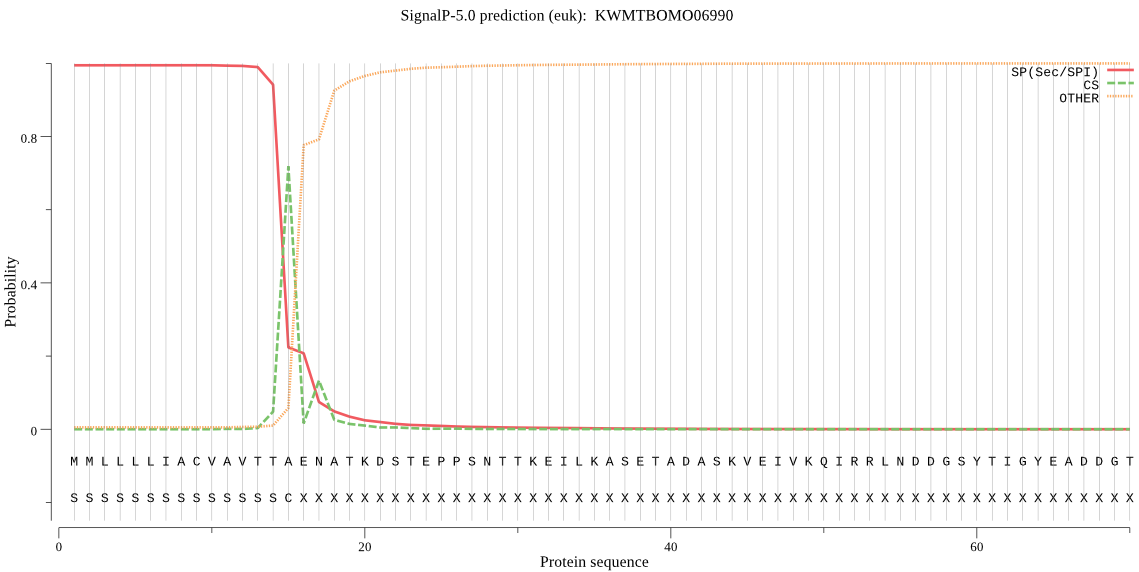
SignalP
Position: 1 - 15,
Likelihood: 0.994643
Length:
655
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00477
Exp number, first 60 AAs:
0.00232
Total prob of N-in:
0.00019
outside
1 - 655
Population Genetic Test Statistics
Pi
135.683932
Theta
161.416332
Tajima's D
-0.48374
CLR
1.116112
CSRT
0.243437828108595
Interpretation
Uncertain
