Gene
KWMTBOMO06986
Pre Gene Modal
BGIBMGA010484
Annotation
PREDICTED:_DDB1-_and_CUL4-associated_factor_7_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 0.923
Sequence
CDS
ATGCCACACAGCAGTTCGAGTACCAGTTCAGGCACGAAACGTAAAGAAATATACAAATACCAAGCGCCATGGCCATTATACTCAATGAATTGGTCTGTCAGACCTGACAAGAGATTCAGGTTAGCCCTGGGCAGTTTTGTTGAGGAATACAACAATAAAGTACAAATAATTTCCCTTGATGAAGACACAAGTGAGTTCACTGCAAAAAGTACCTTTGATCACCCGTACCCGACCACCAAGATTATGTGGATACCTGATAGCAAGGGGGTGTATCCAGACTTGTTGGCAACTAGCGGTGATTATCTACGCATATGGCGTGCCGGAGAGCCGTACACATTATTCGAATGTGTCCTAAATAATAATAAGAATTCTGACTTCTGTGCTCCTCTTACATCCTTTGATTGGAACGAAGTGGATCCTAACCTCATTGGCACCAGTAGTATTGACACGACCTGCACCATCTGGGGCTTGGAGACTGGACAGGTCTTGGGCAGGGTCAATGAAGTGTCTGGGCACGTGAAGACTCAGTTGATCGCTCATGATAAGGAAGTGTACGACATAGCGTTCAGTAGGGCCGGCGGGGGGCGAGATATGTTCGCTTCCGTGGGCGCCGACGGATCCGTCCGGATGTTCGACCTCAGGCATCTAGAACATTCCACCATCATTTATGAGGACCCTCAGCACACCCCGCTGCTCCGCCTGGCCTGGAACAAGCAGGACCCGAACTACCTGGCCACCATCGCCATGGACGCGTGCGAGGTGATCATACTGGACGTGCGCGTGCCCTGCACCCCGGTGGCCAGGCTCAACAACCACAGGGCGTGCGTCAACGGCATCGCCTGGGCGCCGCACAGTTCTTGTCACATCTGCACCGCCGGAGACGACCACCAGGCCCTCATCTGGGACATACAGCAGATGCCGCGCGCCATAGAGGACCCGATCCTCGCCTACACCGCCGCCGAGGGAGAGGTCAACCAGATCCAGTGGGGCGCCACACAACCCGACTGGATCGCCATCTGCTACAACCGGCACGCCGAGATACTGCGCGTCTGA
Protein
MPHSSSSTSSGTKRKEIYKYQAPWPLYSMNWSVRPDKRFRLALGSFVEEYNNKVQIISLDEDTSEFTAKSTFDHPYPTTKIMWIPDSKGVYPDLLATSGDYLRIWRAGEPYTLFECVLNNNKNSDFCAPLTSFDWNEVDPNLIGTSSIDTTCTIWGLETGQVLGRVNEVSGHVKTQLIAHDKEVYDIAFSRAGGGRDMFASVGADGSVRMFDLRHLEHSTIIYEDPQHTPLLRLAWNKQDPNYLATIAMDACEVIILDVRVPCTPVARLNNHRACVNGIAWAPHSSCHICTAGDDHQALIWDIQQMPRAIEDPILAYTAAEGEVNQIQWGATQPDWIAICYNRHAEILRV
Summary
Similarity
Belongs to the class-I aminoacyl-tRNA synthetase family.
Uniprot
A0A3S2PGH9
A0A2W1BTC9
A0A2A4K3T6
A0A2H1W9M2
A0A212EX12
A0A194QPE3
+ More
A0A194QIX9 A0A2Z5U7M8 A0A2J7RNU1 A0A2J7RNU3 A0A2P8YLS6 A0A2J7RNU2 A0A067R119 A0A232F4N0 T1HWD9 A0A154PGF2 A0A0P4VR09 E2BQZ7 A0A195CU17 A0A023F5V4 A0A0L7QTZ2 A0A2A3EE51 A0A088AS01 A0A0V0G9X0 A0A069DRZ7 A0A1Y1L0Z3 A0A026WM51 A0A1B6ERH5 A0A1W4WGR8 A0A0J7KM37 A0A1B6KXQ2 D6X2C7 A0A0M9AAL1 E2AHK0 A0A1B0GAE8 N6TMD4 A0A1L8DAL1 A0A1B0DLI1 A0A151WMK5 A0A0A9YD58 A0A1B0B9P1 A0A195E3L4 A0A182JKY7 A0A158NKW7 A0A1L8DAV0 A0A023ERD7 Q16QR0 A0A2H8TJZ8 J9JTS2 A0A2S2PAH2 A0A2M4CS55 A0A2S2Q3M4 A0A1A9XKN4 A0A182XY71 A0A182VVE9 A0A182LU38 A0A2M4CX96 A0A1B6DRM6 A0A1S4FSU1 A0A182KFY9 A0A182GM34 A0A084VVB6 A0A182RYS3 A0A182PU07 B0WSY7 A0A182QEA9 A0A182USZ9 Q7PRP8 A0A195FTS7 A0A2M4BTQ5 F4WAE3 W5JWV6 A0A182FID2 A0A2M4APR6 A0A2R7VUR6 A0A182X7H2 F5HJU6 A0A182I9W8 B4H2Z9 A0A1A9X3A6 A0A0K8V9Q8 A0A1A9UE49 A0A034WFR4 T1JKQ2 A0A1W4VWR8 B4NUV7 A0A3B0JWV7 Q29JF3 B4PZK0 B3MRG1 B3NY65 A0A0A1XRP9 A0A1B0A2Y0 Q9VR53 A0A1I8N6U4 A0A1I8Q193 B4JWR4 B4L2V4 B4M382 A0A0N8AZA6
A0A194QIX9 A0A2Z5U7M8 A0A2J7RNU1 A0A2J7RNU3 A0A2P8YLS6 A0A2J7RNU2 A0A067R119 A0A232F4N0 T1HWD9 A0A154PGF2 A0A0P4VR09 E2BQZ7 A0A195CU17 A0A023F5V4 A0A0L7QTZ2 A0A2A3EE51 A0A088AS01 A0A0V0G9X0 A0A069DRZ7 A0A1Y1L0Z3 A0A026WM51 A0A1B6ERH5 A0A1W4WGR8 A0A0J7KM37 A0A1B6KXQ2 D6X2C7 A0A0M9AAL1 E2AHK0 A0A1B0GAE8 N6TMD4 A0A1L8DAL1 A0A1B0DLI1 A0A151WMK5 A0A0A9YD58 A0A1B0B9P1 A0A195E3L4 A0A182JKY7 A0A158NKW7 A0A1L8DAV0 A0A023ERD7 Q16QR0 A0A2H8TJZ8 J9JTS2 A0A2S2PAH2 A0A2M4CS55 A0A2S2Q3M4 A0A1A9XKN4 A0A182XY71 A0A182VVE9 A0A182LU38 A0A2M4CX96 A0A1B6DRM6 A0A1S4FSU1 A0A182KFY9 A0A182GM34 A0A084VVB6 A0A182RYS3 A0A182PU07 B0WSY7 A0A182QEA9 A0A182USZ9 Q7PRP8 A0A195FTS7 A0A2M4BTQ5 F4WAE3 W5JWV6 A0A182FID2 A0A2M4APR6 A0A2R7VUR6 A0A182X7H2 F5HJU6 A0A182I9W8 B4H2Z9 A0A1A9X3A6 A0A0K8V9Q8 A0A1A9UE49 A0A034WFR4 T1JKQ2 A0A1W4VWR8 B4NUV7 A0A3B0JWV7 Q29JF3 B4PZK0 B3MRG1 B3NY65 A0A0A1XRP9 A0A1B0A2Y0 Q9VR53 A0A1I8N6U4 A0A1I8Q193 B4JWR4 B4L2V4 B4M382 A0A0N8AZA6
Pubmed
28756777
22118469
26354079
29403074
24845553
28648823
+ More
27129103 20798317 25474469 26334808 28004739 24508170 30249741 18362917 19820115 23537049 25401762 26823975 21347285 24945155 17510324 25244985 26483478 24438588 12364791 21719571 20920257 23761445 14747013 17210077 17994087 25348373 15632085 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 18057021
27129103 20798317 25474469 26334808 28004739 24508170 30249741 18362917 19820115 23537049 25401762 26823975 21347285 24945155 17510324 25244985 26483478 24438588 12364791 21719571 20920257 23761445 14747013 17210077 17994087 25348373 15632085 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 18057021
EMBL
RSAL01000043
RVE50738.1
KZ149906
PZC78309.1
NWSH01000192
PCG78564.1
+ More
ODYU01005429 ODYU01006905 SOQ46262.1 SOQ49174.1 AGBW02011856 OWR46011.1 KQ461194 KPJ06845.1 KQ458671 KPJ05513.1 AP017552 BBB06984.1 NEVH01002147 PNF42506.1 PNF42507.1 PYGN01000503 PSN45205.1 PNF42505.1 KK852819 KDR15607.1 NNAY01000958 OXU25731.1 ACPB03008408 KQ434892 KZC10544.1 GDKW01001640 JAI54955.1 GL449840 EFN81883.1 KQ977306 KYN03629.1 GBBI01002145 JAC16567.1 KQ414736 KOC62118.1 KZ288271 PBC29744.1 GECL01001556 JAP04568.1 GBGD01002056 JAC86833.1 GEZM01067904 GEZM01067903 JAV67333.1 KK107159 QOIP01000001 EZA56741.1 RLU26222.1 GECZ01029259 JAS40510.1 LBMM01005671 KMQ91304.1 GEBQ01025454 GEBQ01023741 JAT14523.1 JAT16236.1 KQ971371 EFA09880.1 KQ435710 KOX79886.1 GL439548 EFN67103.1 CCAG010011418 CCAG010011419 APGK01029540 KB740735 KB631642 ENN79188.1 ERL84905.1 GFDF01010578 JAV03506.1 AJVK01006674 AJVK01006675 AJVK01006676 KQ982937 KYQ49104.1 GBHO01016134 GBRD01012147 GDHC01011080 JAG27470.1 JAG53677.1 JAQ07549.1 JXJN01010502 JXJN01010503 KQ979701 KYN19661.1 ADTU01018978 GFDF01010579 JAV03505.1 GAPW01001853 JAC11745.1 CH477736 EAT36743.1 GFXV01002649 MBW14454.1 ABLF02030005 ABLF02030006 ABLF02030008 ABLF02030010 ABLF02040732 ABLF02040757 ABLF02040768 ABLF02043927 ABLF02063499 GGMR01013842 MBY26461.1 GGFL01003996 MBW68174.1 GGMS01002619 MBY71822.1 AXCM01002613 AXCM01002614 GGFL01005711 MBW69889.1 GEDC01008971 JAS28327.1 JXUM01073760 JXUM01073761 JXUM01073762 JXUM01073763 KQ562794 KXJ75121.1 ATLV01017170 KE525157 KFB41910.1 DS232078 EDS34120.1 AXCN02001335 AAAB01008847 EAA06830.4 KQ981281 KYN43294.1 GGFJ01006997 MBW56138.1 GL888048 EGI68826.1 ADMH02000148 ETN67604.1 GGFK01009465 MBW42786.1 KK854025 PTY09665.1 EGK96562.1 APCN01001063 CH479205 EDW30831.1 GDHF01016722 JAI35592.1 GAKP01004541 JAC54411.1 JH431912 CH984300 EDX16392.1 OUUW01000003 SPP77836.1 CH379063 EAL32348.3 CM000162 EDX03128.1 CH902622 EDV34366.1 CH954180 EDV47650.1 GBXI01000323 JAD13969.1 AE014298 BT099938 AAF50953.2 ACX36514.1 AFH07509.1 AFH07510.1 AHN59996.1 CH916376 EDV95190.1 CH933810 EDW06921.1 KRF93874.1 CH940651 EDW65257.1 KRF82147.1 GDIQ01228211 GDIP01091607 GDIQ01088752 LRGB01002793 JAK23514.1 JAM12108.1 KZS06325.1
ODYU01005429 ODYU01006905 SOQ46262.1 SOQ49174.1 AGBW02011856 OWR46011.1 KQ461194 KPJ06845.1 KQ458671 KPJ05513.1 AP017552 BBB06984.1 NEVH01002147 PNF42506.1 PNF42507.1 PYGN01000503 PSN45205.1 PNF42505.1 KK852819 KDR15607.1 NNAY01000958 OXU25731.1 ACPB03008408 KQ434892 KZC10544.1 GDKW01001640 JAI54955.1 GL449840 EFN81883.1 KQ977306 KYN03629.1 GBBI01002145 JAC16567.1 KQ414736 KOC62118.1 KZ288271 PBC29744.1 GECL01001556 JAP04568.1 GBGD01002056 JAC86833.1 GEZM01067904 GEZM01067903 JAV67333.1 KK107159 QOIP01000001 EZA56741.1 RLU26222.1 GECZ01029259 JAS40510.1 LBMM01005671 KMQ91304.1 GEBQ01025454 GEBQ01023741 JAT14523.1 JAT16236.1 KQ971371 EFA09880.1 KQ435710 KOX79886.1 GL439548 EFN67103.1 CCAG010011418 CCAG010011419 APGK01029540 KB740735 KB631642 ENN79188.1 ERL84905.1 GFDF01010578 JAV03506.1 AJVK01006674 AJVK01006675 AJVK01006676 KQ982937 KYQ49104.1 GBHO01016134 GBRD01012147 GDHC01011080 JAG27470.1 JAG53677.1 JAQ07549.1 JXJN01010502 JXJN01010503 KQ979701 KYN19661.1 ADTU01018978 GFDF01010579 JAV03505.1 GAPW01001853 JAC11745.1 CH477736 EAT36743.1 GFXV01002649 MBW14454.1 ABLF02030005 ABLF02030006 ABLF02030008 ABLF02030010 ABLF02040732 ABLF02040757 ABLF02040768 ABLF02043927 ABLF02063499 GGMR01013842 MBY26461.1 GGFL01003996 MBW68174.1 GGMS01002619 MBY71822.1 AXCM01002613 AXCM01002614 GGFL01005711 MBW69889.1 GEDC01008971 JAS28327.1 JXUM01073760 JXUM01073761 JXUM01073762 JXUM01073763 KQ562794 KXJ75121.1 ATLV01017170 KE525157 KFB41910.1 DS232078 EDS34120.1 AXCN02001335 AAAB01008847 EAA06830.4 KQ981281 KYN43294.1 GGFJ01006997 MBW56138.1 GL888048 EGI68826.1 ADMH02000148 ETN67604.1 GGFK01009465 MBW42786.1 KK854025 PTY09665.1 EGK96562.1 APCN01001063 CH479205 EDW30831.1 GDHF01016722 JAI35592.1 GAKP01004541 JAC54411.1 JH431912 CH984300 EDX16392.1 OUUW01000003 SPP77836.1 CH379063 EAL32348.3 CM000162 EDX03128.1 CH902622 EDV34366.1 CH954180 EDV47650.1 GBXI01000323 JAD13969.1 AE014298 BT099938 AAF50953.2 ACX36514.1 AFH07509.1 AFH07510.1 AHN59996.1 CH916376 EDV95190.1 CH933810 EDW06921.1 KRF93874.1 CH940651 EDW65257.1 KRF82147.1 GDIQ01228211 GDIP01091607 GDIQ01088752 LRGB01002793 JAK23514.1 JAM12108.1 KZS06325.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000053240
UP000053268
UP000235965
+ More
UP000245037 UP000027135 UP000215335 UP000015103 UP000076502 UP000008237 UP000078542 UP000053825 UP000242457 UP000005203 UP000053097 UP000279307 UP000192223 UP000036403 UP000007266 UP000053105 UP000000311 UP000092444 UP000019118 UP000030742 UP000092462 UP000075809 UP000092460 UP000078492 UP000075880 UP000005205 UP000008820 UP000007819 UP000092443 UP000076408 UP000075920 UP000075883 UP000075881 UP000069940 UP000249989 UP000030765 UP000075900 UP000075885 UP000002320 UP000075886 UP000075903 UP000007062 UP000078541 UP000007755 UP000000673 UP000069272 UP000076407 UP000075840 UP000008744 UP000091820 UP000078200 UP000192221 UP000000304 UP000268350 UP000001819 UP000002282 UP000007801 UP000008711 UP000092445 UP000000803 UP000095301 UP000095300 UP000001070 UP000009192 UP000008792 UP000076858
UP000245037 UP000027135 UP000215335 UP000015103 UP000076502 UP000008237 UP000078542 UP000053825 UP000242457 UP000005203 UP000053097 UP000279307 UP000192223 UP000036403 UP000007266 UP000053105 UP000000311 UP000092444 UP000019118 UP000030742 UP000092462 UP000075809 UP000092460 UP000078492 UP000075880 UP000005205 UP000008820 UP000007819 UP000092443 UP000076408 UP000075920 UP000075883 UP000075881 UP000069940 UP000249989 UP000030765 UP000075900 UP000075885 UP000002320 UP000075886 UP000075903 UP000007062 UP000078541 UP000007755 UP000000673 UP000069272 UP000076407 UP000075840 UP000008744 UP000091820 UP000078200 UP000192221 UP000000304 UP000268350 UP000001819 UP000002282 UP000007801 UP000008711 UP000092445 UP000000803 UP000095301 UP000095300 UP000001070 UP000009192 UP000008792 UP000076858
PRIDE
Pfam
Interpro
IPR019775
WD40_repeat_CS
+ More
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR011650 Peptidase_M20_dimer
IPR036264 Bact_exopeptidase_dim_dom
IPR002933 Peptidase_M20
IPR010159 N-acyl_aa_amidohydrolase
IPR032011 DUF4794
IPR001261 ArgE/DapE_CS
IPR014729 Rossmann-like_a/b/a_fold
IPR015413 Methionyl/Leucyl_tRNA_Synth
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR011650 Peptidase_M20_dimer
IPR036264 Bact_exopeptidase_dim_dom
IPR002933 Peptidase_M20
IPR010159 N-acyl_aa_amidohydrolase
IPR032011 DUF4794
IPR001261 ArgE/DapE_CS
IPR014729 Rossmann-like_a/b/a_fold
IPR015413 Methionyl/Leucyl_tRNA_Synth
Gene 3D
ProteinModelPortal
A0A3S2PGH9
A0A2W1BTC9
A0A2A4K3T6
A0A2H1W9M2
A0A212EX12
A0A194QPE3
+ More
A0A194QIX9 A0A2Z5U7M8 A0A2J7RNU1 A0A2J7RNU3 A0A2P8YLS6 A0A2J7RNU2 A0A067R119 A0A232F4N0 T1HWD9 A0A154PGF2 A0A0P4VR09 E2BQZ7 A0A195CU17 A0A023F5V4 A0A0L7QTZ2 A0A2A3EE51 A0A088AS01 A0A0V0G9X0 A0A069DRZ7 A0A1Y1L0Z3 A0A026WM51 A0A1B6ERH5 A0A1W4WGR8 A0A0J7KM37 A0A1B6KXQ2 D6X2C7 A0A0M9AAL1 E2AHK0 A0A1B0GAE8 N6TMD4 A0A1L8DAL1 A0A1B0DLI1 A0A151WMK5 A0A0A9YD58 A0A1B0B9P1 A0A195E3L4 A0A182JKY7 A0A158NKW7 A0A1L8DAV0 A0A023ERD7 Q16QR0 A0A2H8TJZ8 J9JTS2 A0A2S2PAH2 A0A2M4CS55 A0A2S2Q3M4 A0A1A9XKN4 A0A182XY71 A0A182VVE9 A0A182LU38 A0A2M4CX96 A0A1B6DRM6 A0A1S4FSU1 A0A182KFY9 A0A182GM34 A0A084VVB6 A0A182RYS3 A0A182PU07 B0WSY7 A0A182QEA9 A0A182USZ9 Q7PRP8 A0A195FTS7 A0A2M4BTQ5 F4WAE3 W5JWV6 A0A182FID2 A0A2M4APR6 A0A2R7VUR6 A0A182X7H2 F5HJU6 A0A182I9W8 B4H2Z9 A0A1A9X3A6 A0A0K8V9Q8 A0A1A9UE49 A0A034WFR4 T1JKQ2 A0A1W4VWR8 B4NUV7 A0A3B0JWV7 Q29JF3 B4PZK0 B3MRG1 B3NY65 A0A0A1XRP9 A0A1B0A2Y0 Q9VR53 A0A1I8N6U4 A0A1I8Q193 B4JWR4 B4L2V4 B4M382 A0A0N8AZA6
A0A194QIX9 A0A2Z5U7M8 A0A2J7RNU1 A0A2J7RNU3 A0A2P8YLS6 A0A2J7RNU2 A0A067R119 A0A232F4N0 T1HWD9 A0A154PGF2 A0A0P4VR09 E2BQZ7 A0A195CU17 A0A023F5V4 A0A0L7QTZ2 A0A2A3EE51 A0A088AS01 A0A0V0G9X0 A0A069DRZ7 A0A1Y1L0Z3 A0A026WM51 A0A1B6ERH5 A0A1W4WGR8 A0A0J7KM37 A0A1B6KXQ2 D6X2C7 A0A0M9AAL1 E2AHK0 A0A1B0GAE8 N6TMD4 A0A1L8DAL1 A0A1B0DLI1 A0A151WMK5 A0A0A9YD58 A0A1B0B9P1 A0A195E3L4 A0A182JKY7 A0A158NKW7 A0A1L8DAV0 A0A023ERD7 Q16QR0 A0A2H8TJZ8 J9JTS2 A0A2S2PAH2 A0A2M4CS55 A0A2S2Q3M4 A0A1A9XKN4 A0A182XY71 A0A182VVE9 A0A182LU38 A0A2M4CX96 A0A1B6DRM6 A0A1S4FSU1 A0A182KFY9 A0A182GM34 A0A084VVB6 A0A182RYS3 A0A182PU07 B0WSY7 A0A182QEA9 A0A182USZ9 Q7PRP8 A0A195FTS7 A0A2M4BTQ5 F4WAE3 W5JWV6 A0A182FID2 A0A2M4APR6 A0A2R7VUR6 A0A182X7H2 F5HJU6 A0A182I9W8 B4H2Z9 A0A1A9X3A6 A0A0K8V9Q8 A0A1A9UE49 A0A034WFR4 T1JKQ2 A0A1W4VWR8 B4NUV7 A0A3B0JWV7 Q29JF3 B4PZK0 B3MRG1 B3NY65 A0A0A1XRP9 A0A1B0A2Y0 Q9VR53 A0A1I8N6U4 A0A1I8Q193 B4JWR4 B4L2V4 B4M382 A0A0N8AZA6
PDB
4PSX
E-value=1.99465e-06,
Score=123
Ontologies
GO
Topology
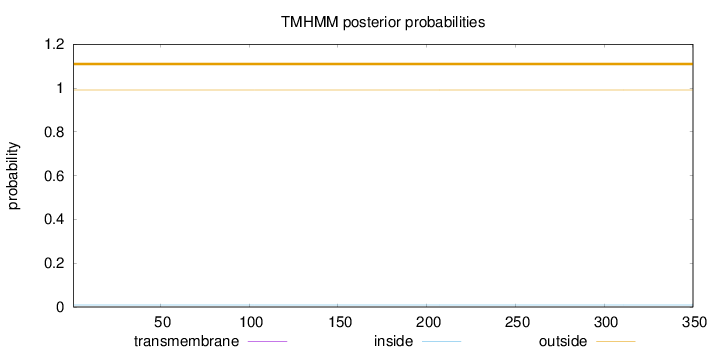
Length:
350
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00154
Exp number, first 60 AAs:
0.00051
Total prob of N-in:
0.00895
outside
1 - 350
Population Genetic Test Statistics
Pi
47.37018
Theta
73.925844
Tajima's D
-1.228133
CLR
0.612149
CSRT
0.100844957752112
Interpretation
Uncertain
