Pre Gene Modal
BGIBMGA010499
Annotation
PREDICTED:_spermatogenesis-associated_protein_5_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.022 Mitochondrial Reliability : 1.656
Sequence
CDS
ATGCCGCCTAAACAAGCTAAGAAAACTCTTTGGGTTGAATGTGAGAAATGTGGATGCAGCGTAACGTCAAACGACAAGAATGTTCACACGGAATTGAATTGTGCAAAAGATCAAGTGAAATGCCCATTTATTTATAACGGAGAGCTAAATACATGGCTCGAAAGGACCGGATCCCCCGCTGATGGAATTCCTAGAGACGCTGTTATGGTACATCCTTCGGCTGGTTCGCTTATAAATGCAGTTATTGGAGGAGCATTACAACTAACCCGCGAGGGTGCACCAGCGATGTTGAAGCGTATGTGGCCCTCCAAAACGTCTTCCCCAGCTGCTGTCATACTTCCTGCCGCTGACATTCCTGCGTCATGGAAAAATGAAAATAAGAAAATTACAGTAGCAGTTCTACAAGAAAATGTAAGTGAAGCCACGGAAGTCATAATAAAAATAAAAAATAACAAAATCAAAACAGACATCTGTGACATATTGAAACATCATTTCTATAAAAATTATGTAGCAGTCGGTACTATACTAGTGTATTATTATTTTGGTAAAAGATTGGAGATAGAAGTGATCACTGTGAAGTCCAAAGATGAAAGTGATAACTCTAGTAAAGACTGGTTTGTTCTCAGTAATGAAACAATTTGGAGTATGAGTAAAGAATCTGAACCAAAAACACAAAAGAAACCAATTTCATTAGGTGGAGTTGATGATATTGTGGAAGAAATCCAGACATACATTAAAATATCCTTTAATGAGGCTGGTTTAAAAGCTAGTTTCCAACCAACTAGAGGATTACTTATTTATGGACATTCAGGGATTGGGAAAACTGCCATCTGTAAATACTTAATTGAGAATTTCAAATGTTTCCATGTAGAAATAAATGGACCAGCAATATTTAGCAAATATTTTGGAGAAACAGAAAACACAATGAAAAGTTTATTTGACAAGGCCGTTAATAATCAACCAAGCATTATATTAGTTGATGAAATTGAAACTATATGTCCCAGGCGCTCATCTGGGAGTACTGAACAAGAGAGACGCATTACTTCAGCCTTTGTATCATTTTTAGACAGTTTACATGATGAAAATTGCAAGGTCTTTGTACTAGCTACAACCAGAAAGCCTGAAGCCATAGATCCTATGTTAAGGAGGTTTGGAAGATTAGACAAAGAAATAGAAATACCGGTTCCAGACAGAGTGCGGAGGAAAGATATCTTGTATAGCTTACTGAAAAATTTACCAAATCAGATGTCTTCGCAAGATATTGAGACAATATCGGATATTGCTCATGGATATGTAGCTGCTGATATAGTGAATCTGTGTACACAAGCTGCCATGAAATGCATAAAGCGTTCAGCTAAAAACATTGAGAAGGAGGACTTGATTGCAGCAGTGACAATAGTCAGACCAAGCGCTATGAGGGAGCTTTTAATAGAAGTACCTAATATTAGATGGGCAGATATTGGTGGTATGGCAGATCTGAAGTTAAAATTACGACAGGCTGTCGAATGGCCATTGAAGCACGCTGAAAGCTTCAAGAGGCTGGGAGTGAAGCCTCCAGGCGGAGTATTGATGTATGGACCTCCAGGTTGCTCTAAAACTATGATAGCTAAAGCTTTAGCAACAGAAAGTGGCCTTAATTTTTTGTCTATTAAAGGTCCTGAACTGTTTTCGAAATGGGTGGGAGAATCTGAGCGGGCTGTCCGAGATTTGTTCACTAAAGCACGCCAAGTGGCACCGTCTGTGATATTTTTTGACGAGATGGATGCCATAGGCGGTGAAAGGGGTGCAGGGGAGGCCGGTGTGCATGAACGAGTGCTGGCACAACTCCTCACTGAGTTAGATGGTGTTGTACCTCTAAATTCTGTGACTATTTTAGCTGCTACCAACAGACCGGACAGGATGGATAGAGCACTACTTCGACCTGGAAGACTAGATCGATTGATCTACGTGCCTTTACCGGACATAGAGACGAGACTACAGATCTTGGAACTGAAGCTTCAAACAATGAGTATTGGTGACGATGTTAACCCTCATGCTTTAGCGTCAAGAACCGAGGGATTTTCTGGAGCCGAGTTGAATGCATTATGTCAGGAAGCTGCTTTGAGGGCTCTAGAAAAAGATCTCGACTGTGCAGAAGTTACTATGACTTATTTCGAGGATGTTTTAAGAAACTTTAAACCCAGAACACCTCATACATTACTGAAAGTCTATGAAGAATTCTGTCTCGGGTAG
Protein
MPPKQAKKTLWVECEKCGCSVTSNDKNVHTELNCAKDQVKCPFIYNGELNTWLERTGSPADGIPRDAVMVHPSAGSLINAVIGGALQLTREGAPAMLKRMWPSKTSSPAAVILPAADIPASWKNENKKITVAVLQENVSEATEVIIKIKNNKIKTDICDILKHHFYKNYVAVGTILVYYYFGKRLEIEVITVKSKDESDNSSKDWFVLSNETIWSMSKESEPKTQKKPISLGGVDDIVEEIQTYIKISFNEAGLKASFQPTRGLLIYGHSGIGKTAICKYLIENFKCFHVEINGPAIFSKYFGETENTMKSLFDKAVNNQPSIILVDEIETICPRRSSGSTEQERRITSAFVSFLDSLHDENCKVFVLATTRKPEAIDPMLRRFGRLDKEIEIPVPDRVRRKDILYSLLKNLPNQMSSQDIETISDIAHGYVAADIVNLCTQAAMKCIKRSAKNIEKEDLIAAVTIVRPSAMRELLIEVPNIRWADIGGMADLKLKLRQAVEWPLKHAESFKRLGVKPPGGVLMYGPPGCSKTMIAKALATESGLNFLSIKGPELFSKWVGESERAVRDLFTKARQVAPSVIFFDEMDAIGGERGAGEAGVHERVLAQLLTELDGVVPLNSVTILAATNRPDRMDRALLRPGRLDRLIYVPLPDIETRLQILELKLQTMSIGDDVNPHALASRTEGFSGAELNALCQEAALRALEKDLDCAEVTMTYFEDVLRNFKPRTPHTLLKVYEEFCLG
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
H9JLU7
A0A2H1VZR4
A0A2A4K480
A0A2W1BZM4
A0A0L7LLW5
A0A194QNB1
+ More
A0A194QJE7 A0A2Z5U7R0 A0A154PCH5 A0A1Y1MJJ3 D2A1M7 A0A087ZYR1 A0A158NRA1 A0A067RK70 A0A2A3EFH5 A0A195FKQ6 E2A0F9 K7J0H4 F4WB09 A0A151IFU5 A0A0C9RD15 A0A232F794 A0A2J7Q8C5 A0A195BFD6 A0A3L8DJT1 A0A026W722 E9IRZ0 A0A0L7RE17 A0A151IUH9 A0A0J7N0I6 A0A151WMZ6 A0A0N0BGV0 A0A1B6KMP5 A0A1W4WU78 A0A146M8T5 A0A023F167 T1I598 A0A0L8GXG6 A0A034VNP9 U4UMH4 A7SBC8 A0A0A1XED8 A0A0L8GX43 A0A146V908 A0A2I4C6X2 A0A1I8NS54 A0A146V935 A0A146ZMZ8 A0A3Q2PQF1 A0A0A1WGD8 A0A1S4FHW4 A0A0A1XJ36 A0A1I8MWB5 W8BTT9 E2C0P7 A0A0L0C8W0 A0A2R5LAJ1 B4MDM1 A0A293MHT1 A0A1Y1VIF0 A0A3Q2EIZ3 V4ART9 A0A0K8UKG2 A0A2D4N8L1 A0A3P9NIY6 M3Z113 A0A2U9C5I1 A0A1Y1Y9L2 A0A372RAN0 A0A2Y9S8F2 A0A3Q1LMS2 A0A1Y1ZVZ5 U6CRX4 M4AVA4 L8IBC4 A0A3B3T2W5 A0A2D4JAP7 A0A383ZBM1 A0A015IBG5 A0A3Q1HKM1 A0A2D0PKY9 A0A2Y9KJM1 G1MGL2 A0A0N8CJZ6 A0A2Y9Q9L3 A0A1Z5KUA8 A0A167MNS9 A0A384BYQ4 U9SHV9 A0A015JVN2 U5EEZ0 A0A3Q7WXE2 A0A3B3UGN3 A0A341BGW1 A0A2U4AR66
A0A194QJE7 A0A2Z5U7R0 A0A154PCH5 A0A1Y1MJJ3 D2A1M7 A0A087ZYR1 A0A158NRA1 A0A067RK70 A0A2A3EFH5 A0A195FKQ6 E2A0F9 K7J0H4 F4WB09 A0A151IFU5 A0A0C9RD15 A0A232F794 A0A2J7Q8C5 A0A195BFD6 A0A3L8DJT1 A0A026W722 E9IRZ0 A0A0L7RE17 A0A151IUH9 A0A0J7N0I6 A0A151WMZ6 A0A0N0BGV0 A0A1B6KMP5 A0A1W4WU78 A0A146M8T5 A0A023F167 T1I598 A0A0L8GXG6 A0A034VNP9 U4UMH4 A7SBC8 A0A0A1XED8 A0A0L8GX43 A0A146V908 A0A2I4C6X2 A0A1I8NS54 A0A146V935 A0A146ZMZ8 A0A3Q2PQF1 A0A0A1WGD8 A0A1S4FHW4 A0A0A1XJ36 A0A1I8MWB5 W8BTT9 E2C0P7 A0A0L0C8W0 A0A2R5LAJ1 B4MDM1 A0A293MHT1 A0A1Y1VIF0 A0A3Q2EIZ3 V4ART9 A0A0K8UKG2 A0A2D4N8L1 A0A3P9NIY6 M3Z113 A0A2U9C5I1 A0A1Y1Y9L2 A0A372RAN0 A0A2Y9S8F2 A0A3Q1LMS2 A0A1Y1ZVZ5 U6CRX4 M4AVA4 L8IBC4 A0A3B3T2W5 A0A2D4JAP7 A0A383ZBM1 A0A015IBG5 A0A3Q1HKM1 A0A2D0PKY9 A0A2Y9KJM1 G1MGL2 A0A0N8CJZ6 A0A2Y9Q9L3 A0A1Z5KUA8 A0A167MNS9 A0A384BYQ4 U9SHV9 A0A015JVN2 U5EEZ0 A0A3Q7WXE2 A0A3B3UGN3 A0A341BGW1 A0A2U4AR66
Pubmed
19121390
28756777
26227816
26354079
28004739
18362917
+ More
19820115 21347285 24845553 20798317 20075255 21719571 28648823 30249741 24508170 21282665 26823975 25474469 25348373 23537049 17615350 25830018 17510324 25315136 24495485 26108605 17994087 23254933 19393038 23542700 22751099 29240929 20010809 28528879 24813606
19820115 21347285 24845553 20798317 20075255 21719571 28648823 30249741 24508170 21282665 26823975 25474469 25348373 23537049 17615350 25830018 17510324 25315136 24495485 26108605 17994087 23254933 19393038 23542700 22751099 29240929 20010809 28528879 24813606
EMBL
BABH01005129
ODYU01005429
SOQ46263.1
NWSH01000192
PCG78563.1
KZ149906
+ More
PZC78310.1 JTDY01000621 KOB76442.1 KQ461194 KPJ06844.1 KQ458671 KPJ05514.1 AP017552 BBB06983.1 KQ434864 KZC09114.1 GEZM01031035 GEZM01031034 JAV85108.1 KQ971338 EFA02694.2 ADTU01023838 KK852424 KDR24192.1 KZ288269 PBC29906.1 KQ981522 KYN40579.1 GL435586 EFN73096.1 GL888056 EGI68629.1 KQ977771 KYM99999.1 GBYB01014484 JAG84251.1 NNAY01000747 OXU26711.1 NEVH01016966 PNF24829.1 KQ976504 KYM82902.1 QOIP01000007 RLU20423.1 KK107390 EZA51421.1 GL765283 EFZ16685.1 KQ414613 KOC69069.1 KQ980970 KYN10935.1 LBMM01012558 KMQ86150.1 KQ982922 KYQ49249.1 KQ435765 KOX75342.1 GEBQ01027248 JAT12729.1 GDHC01002465 JAQ16164.1 GBBI01003946 JAC14766.1 ACPB03000702 KQ420036 KOF81544.1 GAKP01015242 GAKP01015240 GAKP01015238 GAKP01015236 GAKP01015235 JAC43712.1 KB632278 ERL91220.1 DS469615 EDO39006.1 GBXI01009739 GBXI01005439 JAD04553.1 JAD08853.1 KOF81543.1 GCES01072420 JAR13903.1 GCES01072419 JAR13904.1 GCES01018603 JAR67720.1 GBXI01016824 JAC97467.1 GBXI01003271 JAD11021.1 GAMC01003955 GAMC01003953 JAC02603.1 GL451846 EFN78441.1 JRES01000841 KNC27834.1 GGLE01002415 MBY06541.1 CH940661 EDW71282.1 GFWV01014779 MAA39508.1 MCFH01000008 ORX55872.1 KB200869 ESO99957.1 GDHF01025140 JAI27174.1 IACM01157501 LAB41658.1 AEYP01023560 AEYP01023561 AEYP01023562 AEYP01023563 AEYP01023564 AEYP01023565 AEYP01023566 AEYP01023567 AEYP01023568 AEYP01023569 CP026255 AWP11678.1 MCFE01000198 ORX94701.1 QKKE01000156 RGB36039.1 MCOG01000351 ORY14237.1 HAAF01000776 CCP72602.1 JH881568 ELR53518.1 IACK01155118 LAA93572.1 JEMT01029715 EXX51145.1 ACTA01065803 ACTA01073803 ACTA01081803 ACTA01089803 ACTA01097803 ACTA01105803 ACTA01113803 ACTA01121803 ACTA01129803 ACTA01137803 GDIP01124279 JAL79435.1 GFJQ02008365 JAV98604.1 KV440981 OAD73394.1 KI301224 ERZ95473.1 EXX51146.1 GANO01004098 JAB55773.1
PZC78310.1 JTDY01000621 KOB76442.1 KQ461194 KPJ06844.1 KQ458671 KPJ05514.1 AP017552 BBB06983.1 KQ434864 KZC09114.1 GEZM01031035 GEZM01031034 JAV85108.1 KQ971338 EFA02694.2 ADTU01023838 KK852424 KDR24192.1 KZ288269 PBC29906.1 KQ981522 KYN40579.1 GL435586 EFN73096.1 GL888056 EGI68629.1 KQ977771 KYM99999.1 GBYB01014484 JAG84251.1 NNAY01000747 OXU26711.1 NEVH01016966 PNF24829.1 KQ976504 KYM82902.1 QOIP01000007 RLU20423.1 KK107390 EZA51421.1 GL765283 EFZ16685.1 KQ414613 KOC69069.1 KQ980970 KYN10935.1 LBMM01012558 KMQ86150.1 KQ982922 KYQ49249.1 KQ435765 KOX75342.1 GEBQ01027248 JAT12729.1 GDHC01002465 JAQ16164.1 GBBI01003946 JAC14766.1 ACPB03000702 KQ420036 KOF81544.1 GAKP01015242 GAKP01015240 GAKP01015238 GAKP01015236 GAKP01015235 JAC43712.1 KB632278 ERL91220.1 DS469615 EDO39006.1 GBXI01009739 GBXI01005439 JAD04553.1 JAD08853.1 KOF81543.1 GCES01072420 JAR13903.1 GCES01072419 JAR13904.1 GCES01018603 JAR67720.1 GBXI01016824 JAC97467.1 GBXI01003271 JAD11021.1 GAMC01003955 GAMC01003953 JAC02603.1 GL451846 EFN78441.1 JRES01000841 KNC27834.1 GGLE01002415 MBY06541.1 CH940661 EDW71282.1 GFWV01014779 MAA39508.1 MCFH01000008 ORX55872.1 KB200869 ESO99957.1 GDHF01025140 JAI27174.1 IACM01157501 LAB41658.1 AEYP01023560 AEYP01023561 AEYP01023562 AEYP01023563 AEYP01023564 AEYP01023565 AEYP01023566 AEYP01023567 AEYP01023568 AEYP01023569 CP026255 AWP11678.1 MCFE01000198 ORX94701.1 QKKE01000156 RGB36039.1 MCOG01000351 ORY14237.1 HAAF01000776 CCP72602.1 JH881568 ELR53518.1 IACK01155118 LAA93572.1 JEMT01029715 EXX51145.1 ACTA01065803 ACTA01073803 ACTA01081803 ACTA01089803 ACTA01097803 ACTA01105803 ACTA01113803 ACTA01121803 ACTA01129803 ACTA01137803 GDIP01124279 JAL79435.1 GFJQ02008365 JAV98604.1 KV440981 OAD73394.1 KI301224 ERZ95473.1 EXX51146.1 GANO01004098 JAB55773.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000076502
+ More
UP000007266 UP000005203 UP000005205 UP000027135 UP000242457 UP000078541 UP000000311 UP000002358 UP000007755 UP000078542 UP000215335 UP000235965 UP000078540 UP000279307 UP000053097 UP000053825 UP000078492 UP000036403 UP000075809 UP000053105 UP000192223 UP000015103 UP000053454 UP000030742 UP000001593 UP000192220 UP000095300 UP000265000 UP000095301 UP000008237 UP000037069 UP000008792 UP000193719 UP000265020 UP000030746 UP000242638 UP000000715 UP000246464 UP000193498 UP000263633 UP000248484 UP000009136 UP000193920 UP000002852 UP000261540 UP000261681 UP000022910 UP000265040 UP000221080 UP000248482 UP000008912 UP000248483 UP000077315 UP000261680 UP000291021 UP000286642 UP000261500 UP000252040 UP000245320
UP000007266 UP000005203 UP000005205 UP000027135 UP000242457 UP000078541 UP000000311 UP000002358 UP000007755 UP000078542 UP000215335 UP000235965 UP000078540 UP000279307 UP000053097 UP000053825 UP000078492 UP000036403 UP000075809 UP000053105 UP000192223 UP000015103 UP000053454 UP000030742 UP000001593 UP000192220 UP000095300 UP000265000 UP000095301 UP000008237 UP000037069 UP000008792 UP000193719 UP000265020 UP000030746 UP000242638 UP000000715 UP000246464 UP000193498 UP000263633 UP000248484 UP000009136 UP000193920 UP000002852 UP000261540 UP000261681 UP000022910 UP000265040 UP000221080 UP000248482 UP000008912 UP000248483 UP000077315 UP000261680 UP000291021 UP000286642 UP000261500 UP000252040 UP000245320
PRIDE
Interpro
ProteinModelPortal
H9JLU7
A0A2H1VZR4
A0A2A4K480
A0A2W1BZM4
A0A0L7LLW5
A0A194QNB1
+ More
A0A194QJE7 A0A2Z5U7R0 A0A154PCH5 A0A1Y1MJJ3 D2A1M7 A0A087ZYR1 A0A158NRA1 A0A067RK70 A0A2A3EFH5 A0A195FKQ6 E2A0F9 K7J0H4 F4WB09 A0A151IFU5 A0A0C9RD15 A0A232F794 A0A2J7Q8C5 A0A195BFD6 A0A3L8DJT1 A0A026W722 E9IRZ0 A0A0L7RE17 A0A151IUH9 A0A0J7N0I6 A0A151WMZ6 A0A0N0BGV0 A0A1B6KMP5 A0A1W4WU78 A0A146M8T5 A0A023F167 T1I598 A0A0L8GXG6 A0A034VNP9 U4UMH4 A7SBC8 A0A0A1XED8 A0A0L8GX43 A0A146V908 A0A2I4C6X2 A0A1I8NS54 A0A146V935 A0A146ZMZ8 A0A3Q2PQF1 A0A0A1WGD8 A0A1S4FHW4 A0A0A1XJ36 A0A1I8MWB5 W8BTT9 E2C0P7 A0A0L0C8W0 A0A2R5LAJ1 B4MDM1 A0A293MHT1 A0A1Y1VIF0 A0A3Q2EIZ3 V4ART9 A0A0K8UKG2 A0A2D4N8L1 A0A3P9NIY6 M3Z113 A0A2U9C5I1 A0A1Y1Y9L2 A0A372RAN0 A0A2Y9S8F2 A0A3Q1LMS2 A0A1Y1ZVZ5 U6CRX4 M4AVA4 L8IBC4 A0A3B3T2W5 A0A2D4JAP7 A0A383ZBM1 A0A015IBG5 A0A3Q1HKM1 A0A2D0PKY9 A0A2Y9KJM1 G1MGL2 A0A0N8CJZ6 A0A2Y9Q9L3 A0A1Z5KUA8 A0A167MNS9 A0A384BYQ4 U9SHV9 A0A015JVN2 U5EEZ0 A0A3Q7WXE2 A0A3B3UGN3 A0A341BGW1 A0A2U4AR66
A0A194QJE7 A0A2Z5U7R0 A0A154PCH5 A0A1Y1MJJ3 D2A1M7 A0A087ZYR1 A0A158NRA1 A0A067RK70 A0A2A3EFH5 A0A195FKQ6 E2A0F9 K7J0H4 F4WB09 A0A151IFU5 A0A0C9RD15 A0A232F794 A0A2J7Q8C5 A0A195BFD6 A0A3L8DJT1 A0A026W722 E9IRZ0 A0A0L7RE17 A0A151IUH9 A0A0J7N0I6 A0A151WMZ6 A0A0N0BGV0 A0A1B6KMP5 A0A1W4WU78 A0A146M8T5 A0A023F167 T1I598 A0A0L8GXG6 A0A034VNP9 U4UMH4 A7SBC8 A0A0A1XED8 A0A0L8GX43 A0A146V908 A0A2I4C6X2 A0A1I8NS54 A0A146V935 A0A146ZMZ8 A0A3Q2PQF1 A0A0A1WGD8 A0A1S4FHW4 A0A0A1XJ36 A0A1I8MWB5 W8BTT9 E2C0P7 A0A0L0C8W0 A0A2R5LAJ1 B4MDM1 A0A293MHT1 A0A1Y1VIF0 A0A3Q2EIZ3 V4ART9 A0A0K8UKG2 A0A2D4N8L1 A0A3P9NIY6 M3Z113 A0A2U9C5I1 A0A1Y1Y9L2 A0A372RAN0 A0A2Y9S8F2 A0A3Q1LMS2 A0A1Y1ZVZ5 U6CRX4 M4AVA4 L8IBC4 A0A3B3T2W5 A0A2D4JAP7 A0A383ZBM1 A0A015IBG5 A0A3Q1HKM1 A0A2D0PKY9 A0A2Y9KJM1 G1MGL2 A0A0N8CJZ6 A0A2Y9Q9L3 A0A1Z5KUA8 A0A167MNS9 A0A384BYQ4 U9SHV9 A0A015JVN2 U5EEZ0 A0A3Q7WXE2 A0A3B3UGN3 A0A341BGW1 A0A2U4AR66
PDB
5VCA
E-value=7.6167e-120,
Score=1104
Ontologies
GO
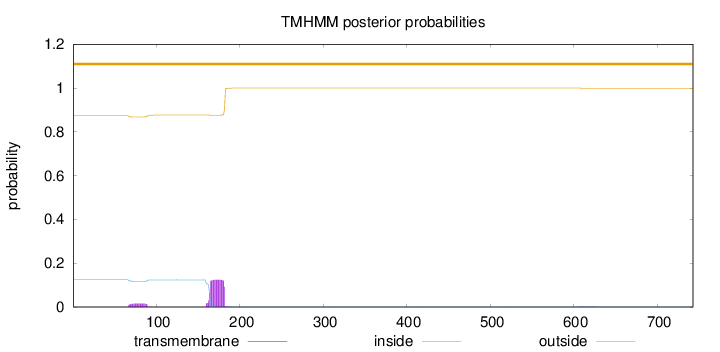
Topology
Length:
743
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.67161
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12535
outside
1 - 743
Population Genetic Test Statistics
Pi
207.76125
Theta
205.0235
Tajima's D
-0.976812
CLR
0.001629
CSRT
0.144442777861107
Interpretation
Uncertain
