Gene
KWMTBOMO06977 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014087
Annotation
peroxiredoxin_[Bombyx_mori]
Full name
Peroxiredoxin
Location in the cell
Cytoplasmic Reliability : 1.304 Extracellular Reliability : 1.343
Sequence
CDS
ATGGCACCAATTAAGGTCGGGGATCAGCTGCCTGCTGCTGATTTATTCGAAGATTCTCCCGCTAATAAGGTGAATATTTGTGAGTTGACGGCGGGAAAGAAGGTTGTATTATTTGCGGTGCCGGGCGCCTTCACCCCGGGATGTTCTAAAACACACTTGCCGGGATACGTACAGAACGCAGATAAACTGAAATCGGATGGAGTTGCTGAAATAGTGTGCGTGTCTGTTAATGACCCGTATGTGATGGCGGCTTGGGGAGCTCAGCACAACACTAAAGGAAAGGTGCGTATGCTAGCCGATCCCAGCGGCAACTTCATCAAGGCTCTGGACCTGGGCACCAATCTGCCGCCGCTCGGAGGTTTCCGCTCCAAAAGGTTCTCGATGGTCATCGTTGACAGCAAGGTCCAAGATCTGAATGTGGAGCCCGATGGCACTGGCCTGTCTTGTTCTCTCGCCGATAAGATCAAAGTGAAGTAG
Protein
MAPIKVGDQLPAADLFEDSPANKVNICELTAGKKVVLFAVPGAFTPGCSKTHLPGYVQNADKLKSDGVAEIVCVSVNDPYVMAAWGAQHNTKGKVRMLADPSGNFIKALDLGTNLPPLGGFRSKRFSMVIVDSKVQDLNVEPDGTGLSCSLADKIKVK
Summary
Description
Thiol-specific peroxidase that catalyzes the reduction of hydrogen peroxide and organic hydroperoxides to water and alcohols, respectively. Plays a role in cell protection against oxidative stress by detoxifying peroxides.
Catalytic Activity
[protein]-dithiol + a hydroperoxide = [protein]-disulfide + an alcohol + H2O
Similarity
Belongs to the peroxiredoxin family. Prx5 subfamily.
Uniprot
Q1HQ45
H9JX22
A0A194QPV0
S4P1X4
A0A212F1Z5
I4DJY2
+ More
A0A194QPD3 I4DN44 A0A2S0RQQ2 A0A3G1T1G6 A0A2A4JQH8 A0A2W1B8Y3 A0A2H1VLR7 A0A068LE92 A0A0L7L6R3 W8CE18 T1DIT5 A0A2M4CSI4 A0A2M4C0G9 A0A2M3Z451 A0A2M3Z481 A0A2M4A3W2 A0A2C9GH06 A0A0A1XIS4 A0A2M3Z505 T1DN66 T1D4Z0 A0A087ZJU1 F4YSY7 Q5UG08 A0A023EHM5 Q173J1 A0A182GU91 A0A023EJ85 A0A0C9Q5Z5 A0A182P9L5 A0A182MPI7 A0A034V1V6 A0A0K8V2N3 A0A182Q7B9 A0A182UES7 A0A182RNL3 A0A182X9M3 A0A3F2YUT3 A0A182HKK7 A0A182IKM5 A0NH65 A0A182YBC5 A0A1Q3G410 A0A182V9T0 W5J4A6 A0A182TN34 B0WM11 A0A1B0D6A3 A0A182S7J3 A0A182NQJ0 A0A182K595 A0A224XLM6 A0A1Y1LXP9 A8CA01 A0A023EIV1 A0A1L8DU06 A0A023F8A5 A0A1I8NES2 T1P7W5 A0A1I8NQW7 A0A161MH80 A0A1L8EEU4 A0A0V0G2U3 F1AG56 A0A0K8TSR3 U5EPT3 A0A0A9YT48 A0A0K8SP46 A0A1A9XWA4 A0A1B0FCN0 A0A0A9YQP0 A0A0K8SQ33 A0A1L7LQG2 Q694A8 A0A067RJ59 K7J041 A0A1A9WJL9 A0A1A9VFL3 A0A154P4C3 B4JUT0 B4LZ87 B4PL69 B4QV16 B3P0D6 Q6XHE3 B4IBA3 A0A1W4UK91 R4G2Z2 A0A1J1I9M5 A0A232EV52 B5DX88 B4GLK9 Q960M4 L0AUL0
A0A194QPD3 I4DN44 A0A2S0RQQ2 A0A3G1T1G6 A0A2A4JQH8 A0A2W1B8Y3 A0A2H1VLR7 A0A068LE92 A0A0L7L6R3 W8CE18 T1DIT5 A0A2M4CSI4 A0A2M4C0G9 A0A2M3Z451 A0A2M3Z481 A0A2M4A3W2 A0A2C9GH06 A0A0A1XIS4 A0A2M3Z505 T1DN66 T1D4Z0 A0A087ZJU1 F4YSY7 Q5UG08 A0A023EHM5 Q173J1 A0A182GU91 A0A023EJ85 A0A0C9Q5Z5 A0A182P9L5 A0A182MPI7 A0A034V1V6 A0A0K8V2N3 A0A182Q7B9 A0A182UES7 A0A182RNL3 A0A182X9M3 A0A3F2YUT3 A0A182HKK7 A0A182IKM5 A0NH65 A0A182YBC5 A0A1Q3G410 A0A182V9T0 W5J4A6 A0A182TN34 B0WM11 A0A1B0D6A3 A0A182S7J3 A0A182NQJ0 A0A182K595 A0A224XLM6 A0A1Y1LXP9 A8CA01 A0A023EIV1 A0A1L8DU06 A0A023F8A5 A0A1I8NES2 T1P7W5 A0A1I8NQW7 A0A161MH80 A0A1L8EEU4 A0A0V0G2U3 F1AG56 A0A0K8TSR3 U5EPT3 A0A0A9YT48 A0A0K8SP46 A0A1A9XWA4 A0A1B0FCN0 A0A0A9YQP0 A0A0K8SQ33 A0A1L7LQG2 Q694A8 A0A067RJ59 K7J041 A0A1A9WJL9 A0A1A9VFL3 A0A154P4C3 B4JUT0 B4LZ87 B4PL69 B4QV16 B3P0D6 Q6XHE3 B4IBA3 A0A1W4UK91 R4G2Z2 A0A1J1I9M5 A0A232EV52 B5DX88 B4GLK9 Q960M4 L0AUL0
EC Number
1.11.1.15
Pubmed
19121390
26354079
23622113
22118469
22651552
28756777
+ More
24998343 26227816 24495485 24330624 25830018 20920257 21569254 24945155 17510324 26483478 25348373 20966253 12364791 14747013 17210077 25244985 28004739 17760985 25474469 25315136 26369729 25401762 26823975 26760975 28076409 16164604 24845553 20075255 17994087 17550304 14525923 28648823 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
24998343 26227816 24495485 24330624 25830018 20920257 21569254 24945155 17510324 26483478 25348373 20966253 12364791 14747013 17210077 25244985 28004739 17760985 25474469 25315136 26369729 25401762 26823975 26760975 28076409 16164604 24845553 20075255 17994087 17550304 14525923 28648823 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
DQ443207
ABF51296.1
BABH01042182
KQ458671
KPJ05521.1
GAIX01012275
+ More
JAA80285.1 AGBW02010785 OWR47749.1 AK401600 BAM18222.1 KQ461194 KPJ06835.1 AK402712 BAM19334.1 MF979109 AWA45972.1 MG992390 AXY94828.1 NWSH01000849 PCG73854.1 KZ150222 PZC72092.1 ODYU01003254 SOQ41763.1 KJ701601 AIE44746.1 JTDY01002583 KOB71158.1 GAMC01000971 JAC05585.1 GALA01000771 JAA94081.1 GGFL01004095 MBW68273.1 GGFJ01009613 MBW58754.1 GGFM01002551 MBW23302.1 GGFM01002580 MBW23331.1 GGFK01002166 MBW35487.1 GBXI01003023 JAD11269.1 GGFM01002860 MBW23611.1 GAMD01003150 JAA98440.1 GALA01000768 JAA94084.1 GGFL01004099 MBW68277.1 HM119220 ADH94060.1 AY738253 AAV48533.1 GAPW01004715 GAPW01004712 GAPW01004711 JAC08883.1 CH477423 EAT41205.1 JXUM01088820 JXUM01088821 JXUM01088822 JXUM01088823 JXUM01088824 KQ563727 KXJ73433.1 GAPW01004714 JAC08884.1 GBYB01009548 JAG79315.1 AXCM01000656 GAKP01023415 JAC35543.1 GDHF01033577 GDHF01022772 GDHF01019246 JAI18737.1 JAI29542.1 JAI33068.1 AXCN02001519 APCN01000836 AAAB01008987 EAU75715.2 GFDL01000500 JAV34545.1 ADMH02002125 ETN58721.1 DS231993 EDS30825.1 AJVK01025851 GFTR01003051 JAW13375.1 GEZM01049832 JAV75817.1 EU035825 ABV44727.1 GAPW01004713 JAC08885.1 GFDF01004155 JAV09929.1 GBBI01001156 JAC17556.1 KA644614 AFP59243.1 GEMB01002520 JAS00666.1 GFDG01001589 JAV17210.1 GECL01003673 JAP02451.1 GU582134 ADX36414.1 GDAI01000435 JAI17168.1 GANO01003585 JAB56286.1 GBHO01008235 JAG35369.1 GBRD01010917 GBRD01010915 GDHC01005880 JAG54909.1 JAQ12749.1 CCAG010010183 GBHO01008237 JAG35367.1 GBRD01010914 GBRD01010912 JAG54910.1 FX983168 BAV93786.1 AY625504 AAT85821.1 KK852442 KDR23852.1 KQ434813 KZC06799.1 CH916374 EDV91250.1 CH940650 EDW67094.1 CM000160 EDW95851.1 CM000364 EDX12499.1 CH954181 EDV48651.1 AY232240 AAR10263.1 CH480827 EDW44661.1 ACPB03003651 GAHY01002156 JAA75354.1 CVRI01000042 CRK95670.1 NNAY01002054 OXU22226.1 CM000070 EDY68620.1 CH479185 EDW38433.1 AE014297 AY051983 BT003526 AAF55497.2 AAK93407.1 AAO39530.1 JX915907 AFZ78682.1
JAA80285.1 AGBW02010785 OWR47749.1 AK401600 BAM18222.1 KQ461194 KPJ06835.1 AK402712 BAM19334.1 MF979109 AWA45972.1 MG992390 AXY94828.1 NWSH01000849 PCG73854.1 KZ150222 PZC72092.1 ODYU01003254 SOQ41763.1 KJ701601 AIE44746.1 JTDY01002583 KOB71158.1 GAMC01000971 JAC05585.1 GALA01000771 JAA94081.1 GGFL01004095 MBW68273.1 GGFJ01009613 MBW58754.1 GGFM01002551 MBW23302.1 GGFM01002580 MBW23331.1 GGFK01002166 MBW35487.1 GBXI01003023 JAD11269.1 GGFM01002860 MBW23611.1 GAMD01003150 JAA98440.1 GALA01000768 JAA94084.1 GGFL01004099 MBW68277.1 HM119220 ADH94060.1 AY738253 AAV48533.1 GAPW01004715 GAPW01004712 GAPW01004711 JAC08883.1 CH477423 EAT41205.1 JXUM01088820 JXUM01088821 JXUM01088822 JXUM01088823 JXUM01088824 KQ563727 KXJ73433.1 GAPW01004714 JAC08884.1 GBYB01009548 JAG79315.1 AXCM01000656 GAKP01023415 JAC35543.1 GDHF01033577 GDHF01022772 GDHF01019246 JAI18737.1 JAI29542.1 JAI33068.1 AXCN02001519 APCN01000836 AAAB01008987 EAU75715.2 GFDL01000500 JAV34545.1 ADMH02002125 ETN58721.1 DS231993 EDS30825.1 AJVK01025851 GFTR01003051 JAW13375.1 GEZM01049832 JAV75817.1 EU035825 ABV44727.1 GAPW01004713 JAC08885.1 GFDF01004155 JAV09929.1 GBBI01001156 JAC17556.1 KA644614 AFP59243.1 GEMB01002520 JAS00666.1 GFDG01001589 JAV17210.1 GECL01003673 JAP02451.1 GU582134 ADX36414.1 GDAI01000435 JAI17168.1 GANO01003585 JAB56286.1 GBHO01008235 JAG35369.1 GBRD01010917 GBRD01010915 GDHC01005880 JAG54909.1 JAQ12749.1 CCAG010010183 GBHO01008237 JAG35367.1 GBRD01010914 GBRD01010912 JAG54910.1 FX983168 BAV93786.1 AY625504 AAT85821.1 KK852442 KDR23852.1 KQ434813 KZC06799.1 CH916374 EDV91250.1 CH940650 EDW67094.1 CM000160 EDW95851.1 CM000364 EDX12499.1 CH954181 EDV48651.1 AY232240 AAR10263.1 CH480827 EDW44661.1 ACPB03003651 GAHY01002156 JAA75354.1 CVRI01000042 CRK95670.1 NNAY01002054 OXU22226.1 CM000070 EDY68620.1 CH479185 EDW38433.1 AE014297 AY051983 BT003526 AAF55497.2 AAK93407.1 AAO39530.1 JX915907 AFZ78682.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000218220
UP000037510
+ More
UP000069272 UP000008820 UP000069940 UP000249989 UP000075885 UP000075883 UP000075886 UP000075902 UP000075900 UP000076407 UP000075882 UP000075840 UP000075880 UP000007062 UP000076408 UP000075903 UP000000673 UP000002320 UP000092462 UP000075901 UP000075884 UP000075881 UP000095301 UP000095300 UP000092443 UP000092444 UP000027135 UP000002358 UP000091820 UP000078200 UP000076502 UP000001070 UP000008792 UP000002282 UP000000304 UP000008711 UP000001292 UP000192221 UP000015103 UP000183832 UP000215335 UP000001819 UP000008744 UP000000803
UP000069272 UP000008820 UP000069940 UP000249989 UP000075885 UP000075883 UP000075886 UP000075902 UP000075900 UP000076407 UP000075882 UP000075840 UP000075880 UP000007062 UP000076408 UP000075903 UP000000673 UP000002320 UP000092462 UP000075901 UP000075884 UP000075881 UP000095301 UP000095300 UP000092443 UP000092444 UP000027135 UP000002358 UP000091820 UP000078200 UP000076502 UP000001070 UP000008792 UP000002282 UP000000304 UP000008711 UP000001292 UP000192221 UP000015103 UP000183832 UP000215335 UP000001819 UP000008744 UP000000803
Interpro
Gene 3D
ProteinModelPortal
Q1HQ45
H9JX22
A0A194QPV0
S4P1X4
A0A212F1Z5
I4DJY2
+ More
A0A194QPD3 I4DN44 A0A2S0RQQ2 A0A3G1T1G6 A0A2A4JQH8 A0A2W1B8Y3 A0A2H1VLR7 A0A068LE92 A0A0L7L6R3 W8CE18 T1DIT5 A0A2M4CSI4 A0A2M4C0G9 A0A2M3Z451 A0A2M3Z481 A0A2M4A3W2 A0A2C9GH06 A0A0A1XIS4 A0A2M3Z505 T1DN66 T1D4Z0 A0A087ZJU1 F4YSY7 Q5UG08 A0A023EHM5 Q173J1 A0A182GU91 A0A023EJ85 A0A0C9Q5Z5 A0A182P9L5 A0A182MPI7 A0A034V1V6 A0A0K8V2N3 A0A182Q7B9 A0A182UES7 A0A182RNL3 A0A182X9M3 A0A3F2YUT3 A0A182HKK7 A0A182IKM5 A0NH65 A0A182YBC5 A0A1Q3G410 A0A182V9T0 W5J4A6 A0A182TN34 B0WM11 A0A1B0D6A3 A0A182S7J3 A0A182NQJ0 A0A182K595 A0A224XLM6 A0A1Y1LXP9 A8CA01 A0A023EIV1 A0A1L8DU06 A0A023F8A5 A0A1I8NES2 T1P7W5 A0A1I8NQW7 A0A161MH80 A0A1L8EEU4 A0A0V0G2U3 F1AG56 A0A0K8TSR3 U5EPT3 A0A0A9YT48 A0A0K8SP46 A0A1A9XWA4 A0A1B0FCN0 A0A0A9YQP0 A0A0K8SQ33 A0A1L7LQG2 Q694A8 A0A067RJ59 K7J041 A0A1A9WJL9 A0A1A9VFL3 A0A154P4C3 B4JUT0 B4LZ87 B4PL69 B4QV16 B3P0D6 Q6XHE3 B4IBA3 A0A1W4UK91 R4G2Z2 A0A1J1I9M5 A0A232EV52 B5DX88 B4GLK9 Q960M4 L0AUL0
A0A194QPD3 I4DN44 A0A2S0RQQ2 A0A3G1T1G6 A0A2A4JQH8 A0A2W1B8Y3 A0A2H1VLR7 A0A068LE92 A0A0L7L6R3 W8CE18 T1DIT5 A0A2M4CSI4 A0A2M4C0G9 A0A2M3Z451 A0A2M3Z481 A0A2M4A3W2 A0A2C9GH06 A0A0A1XIS4 A0A2M3Z505 T1DN66 T1D4Z0 A0A087ZJU1 F4YSY7 Q5UG08 A0A023EHM5 Q173J1 A0A182GU91 A0A023EJ85 A0A0C9Q5Z5 A0A182P9L5 A0A182MPI7 A0A034V1V6 A0A0K8V2N3 A0A182Q7B9 A0A182UES7 A0A182RNL3 A0A182X9M3 A0A3F2YUT3 A0A182HKK7 A0A182IKM5 A0NH65 A0A182YBC5 A0A1Q3G410 A0A182V9T0 W5J4A6 A0A182TN34 B0WM11 A0A1B0D6A3 A0A182S7J3 A0A182NQJ0 A0A182K595 A0A224XLM6 A0A1Y1LXP9 A8CA01 A0A023EIV1 A0A1L8DU06 A0A023F8A5 A0A1I8NES2 T1P7W5 A0A1I8NQW7 A0A161MH80 A0A1L8EEU4 A0A0V0G2U3 F1AG56 A0A0K8TSR3 U5EPT3 A0A0A9YT48 A0A0K8SP46 A0A1A9XWA4 A0A1B0FCN0 A0A0A9YQP0 A0A0K8SQ33 A0A1L7LQG2 Q694A8 A0A067RJ59 K7J041 A0A1A9WJL9 A0A1A9VFL3 A0A154P4C3 B4JUT0 B4LZ87 B4PL69 B4QV16 B3P0D6 Q6XHE3 B4IBA3 A0A1W4UK91 R4G2Z2 A0A1J1I9M5 A0A232EV52 B5DX88 B4GLK9 Q960M4 L0AUL0
PDB
3MNG
E-value=4.34304e-46,
Score=460
Ontologies
KEGG
GO
PANTHER
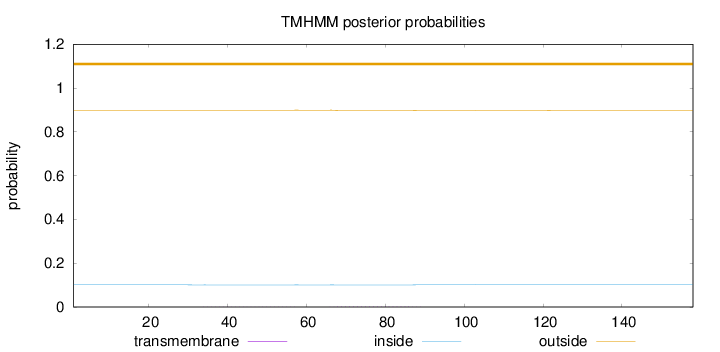
Topology
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05454
Exp number, first 60 AAs:
0.02317
Total prob of N-in:
0.10110
outside
1 - 158
Population Genetic Test Statistics
Pi
165.316921
Theta
171.190931
Tajima's D
-0.178883
CLR
0.493201
CSRT
0.325033748312584
Interpretation
Uncertain
