Gene
KWMTBOMO06976 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008825
Annotation
PREDICTED:_importin-13_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 1.753
Sequence
CDS
ATGGATGCTCCTTCAATGGACACTGTTTACCAGGCGATCAGCGCTCTGTACGATGGCCCAAATGGAAATGAAATTGAGAAGGCTTCGCTATGGCTCGCCGATGTACAGAAATCCATCCACTCATGGAAGATTGCCGACGAGTTGTTGCAGCAAAAAAAGGACTTCAATTCCTGTTATTTCGCGGCACAAACGATGAGAGCGAAAATTCAGCACAACCTTTCAGAGCTACCAGCAGAATCTCATGTACCACTTCGTGACACTCTGATAGCTCACCTGGAGCGCACGACTCCGGACACCACATCTTCAATTCTCACGCAGCTTAGCTTGGCATTGGCGGATCTTGCCCTCCAAATGCCATCTTGGCAAGACTGCATAAGTGACCTCATAAAATTATTTTCAAACAAAGAATATCTACTATTGGAGATTTTGACATTACTACCTCAGGAAATTGACTCTTCGAATTTAAAATTGGGTGAAAATCGCAGAGAAGACATAAAATCCCAACTACGTTTGAATTCACAGACAGTAATGTTATTTCTTAAGCAATGTCTGAATAATTCACAAGGCAGTACAAATGTGGGCTTAAAATCTATAAAGTGCATGACATCATGGCTGCAAGTCCGAGCCGTGAACTTACATGAGATACCCCAGAACACTGTCATAGGATATAGTTTACAAGTTTTGAGAGACCACAATTCTATCAACACATTGCATGATGCTTCTTCTGATTGTATATGTGCACTATTACAATGCTTGGAAGAAGTTAATGACCATGAAAATATAGACAGATTGCTATTTGACAGTGTGGTGGCATTAGAAGAAAGCTACCACTTGGCTGTTGCTCATGAGGAAGAGGAAAAAGTGTCCAATTACGCCAGAGTGTTCACAGAACTAGCTGAATCATTTCTGATGAAAATAATTGATTGTACTGATAATGGGAATACACATTTTGCAATGAAATCATTAGAATTAGCTTTAGTTTGTGTTGGATATCATGACTATGAGGTGGCAGAAATAACATTTAATTTATGGTACAGATTATCAGAGGAAGTGTACAGGAGAGATCACCAACCATTAACAGATGCATTTAAACCTCATATAGAGAGGTTGATCGAAGCTTTGGCAAGGCATTGTCAATGTGAACCTGACTTGACACAATTGCCCGACGAAACTGATGATTTTTATGAATTCCGTCAAAAAGTGATGAGTCTGATCAAGGATGTGGTGTTCATAGTGGGATCAAGTTCAGTTTTCAGACAAATGTTTGCTGCATTACAATCTAACATTTCATGGGAACAGACAGAAGCTGCTCTCTTTGTTATGCAATCTGTTGCTAGAAATATCTTACCGGATGAATACGAATACGTGCCGAAGGTGGTGGAAGCGATACTGTCGATGCCCTCCAGCAGCCACCTGGGCGTGCGCGCCACGTGCATCCTGCTGCTGGGCGAGCTGGCGGACTGGGTGGAGCGGCACGCGGCGTCGTGCCTGGCGCCCTGCGTGGCCATGCTGCTGGCCGCCCTGCCGCAGCCCCGGCTGGCGCTGCCCGCCGCCACCGCGCTGCAGAACATCTGCAAGGCGTGTCCCGGCGGAGCGGCCGAGCACATCGGCAAGCTGCTGGGCGCCGTGCAGGCGCTGGACCAGCTCGCCGCGCCGCCGCCCGCCGCCGCCGCGCTCATGCGGGCCGTCGCGCCCGTCGTGGCCAGCCTGCCGCCCGACCAGATGAGCGCAGCCCTCTGCGAAGCGTCAGCCCCCCAACTGGAGGGTCTCCGTCGGCTGTGCTCGGCTCGGCAGCCCATCAAGCGCGGCACCACGTCGGACCCCGTGCTGTACCTCGACCGGCTCGCCGCGCTGTACCGCGACCTCACGCTCTCCGCCGACCTCAAAAACGTCGCGGCGTCCGAAACGCACCCGTGTCTGCCGGCGCTCACCGAGGCGTGGCCCGTGCTCATCGACGTCATGGACAAGTTCGGCGGCGAGGAGCGCGTGATGGAGCGCGTGTGCCGGGCGGCGCGGTTCGGCGTGCGGGCGGCGCGCGGGGCGGGCGCGGCGCTGCTGCCGGCGCTGTGCGGCGCGCTGGCGCAGCTGTACCCGCGGCACGCGCACTCGTGCGTGCTGTACCTGTGCTCCGTGCTGGTGGACGAGCTGGCGGCGCGGCCGGGCTGCGCGGCGGCGCTGGCGGGGCTGCTGGCCGACCTGCTGCCGCCAGCCTTCGCGCTGCTGCAGCGGGACCACGGCTTCAGGGACCACCCCGACACCGTCGACGATCTGTTCCGGCTCTTCATCAGGTTCCTGCAGCGCATGCCGGTGCAGCTGCTGTCGTGCGGCGTGATGGGCAGCATCCTGCAGTGCGCGTGCTCCGCCGCCGCGCTCGACCACCGCGACGCCAGCGCCTCGCTCATGAACTTCCTCGCGGACTTCGCGCGCCTCGCCAACACCGCGCACCCCGACACCGCCGAGGCGCAGCACGCCAAGGCGCTGGCCGAGGAGCTGGTGGTGACGTACGGCGGCGCGCTGACGGCGGCCGTGCTGGAGGCCAGCGTGCTGCACCTGCACCGCGGGCTCGTGCCCGGCGCCGCCGACGTGCTGCTGGAGCTGGCGCGCGCGGAGAGACGGCACGGCGCCGCGTGGCTGGGGGCCGCCGTGCGCCAGCTGCCCGCGCGCCACACCGCCGCCGCCACGCTGCTGCAGGCGCAGCACTTCCACAACACGGCGCGCAGGTCGGTCCCCCGCAGCGCGTTCGTGCCAAACTTCTTTAGAATCGTTGTGATTATAAACTCGATGTAA
Protein
MDAPSMDTVYQAISALYDGPNGNEIEKASLWLADVQKSIHSWKIADELLQQKKDFNSCYFAAQTMRAKIQHNLSELPAESHVPLRDTLIAHLERTTPDTTSSILTQLSLALADLALQMPSWQDCISDLIKLFSNKEYLLLEILTLLPQEIDSSNLKLGENRREDIKSQLRLNSQTVMLFLKQCLNNSQGSTNVGLKSIKCMTSWLQVRAVNLHEIPQNTVIGYSLQVLRDHNSINTLHDASSDCICALLQCLEEVNDHENIDRLLFDSVVALEESYHLAVAHEEEEKVSNYARVFTELAESFLMKIIDCTDNGNTHFAMKSLELALVCVGYHDYEVAEITFNLWYRLSEEVYRRDHQPLTDAFKPHIERLIEALARHCQCEPDLTQLPDETDDFYEFRQKVMSLIKDVVFIVGSSSVFRQMFAALQSNISWEQTEAALFVMQSVARNILPDEYEYVPKVVEAILSMPSSSHLGVRATCILLLGELADWVERHAASCLAPCVAMLLAALPQPRLALPAATALQNICKACPGGAAEHIGKLLGAVQALDQLAAPPPAAAALMRAVAPVVASLPPDQMSAALCEASAPQLEGLRRLCSARQPIKRGTTSDPVLYLDRLAALYRDLTLSADLKNVAASETHPCLPALTEAWPVLIDVMDKFGGEERVMERVCRAARFGVRAARGAGAALLPALCGALAQLYPRHAHSCVLYLCSVLVDELAARPGCAAALAGLLADLLPPAFALLQRDHGFRDHPDTVDDLFRLFIRFLQRMPVQLLSCGVMGSILQCACSAAALDHRDASASLMNFLADFARLANTAHPDTAEAQHAKALAEELVVTYGGALTAAVLEASVLHLHRGLVPGAADVLLELARAERRHGAAWLGAAVRQLPARHTAAATLLQAQHFHNTARRSVPRSAFVPNFFRIVVIINSM
Summary
Uniprot
A0A2A4JQY8
A0A2W1BCF1
A0A3S2P2K2
A0A212F1Y2
A0A2J7RC24
A0A067R215
+ More
A0A1B6LXV9 A0A0T6AX32 A0A1B6FRA0 A0A023F2N1 A0A0A9ZA41 A0A0K8SR47 A0A1B6DC68 A0A1B6I755 T1I4V9 D2A1S2 A0A154PB99 A0A1Y1N445 A0A0C9R8X6 K7IQL9 A0A232FHY9 A0A1W4XUS1 A0A026W543 E2ACK2 E2B5G1 A0A2A3EF25 E0VRR5 A0A0M9A2Y2 A0A151JME7 A0A0L7RE98 A0A158NXU8 Q17F50 A0A1S3DGY8 A0A1I8MGA5 A0A182GE21 A0A182H7T2 A0A182M6K7 A0A0P6ITY6 A0A1Q3F9Q7 A0A1Q3F9K5 A0A182VQ52 B0WT06 A0A151HY19 A0A182K3M2 A0A195FYK9 A0A131Y0D2 A0A0L8HPQ8 A0A182PS22 A0A2T7P5Z6 A0A0L0CFK2 A0A182VJJ5 Q5TT59 A0A182U4N2 A0A182Y7D7 A0A182KXI0 A0A0K8UQ20 A0A0A1WFH3 A0A182T4N5 A0A1Q3F9P6 A0A1Q3F9K1 A0A182Q5Y4 B4N0H1 A0A084VYU2 A0A182NLH0 A0A1I8P2Y7 A0A182FT12 A0A182RZG6 Q29KW7 B4GSP8 A0A2C9H8L6 A0A1S4GJ51 A0A182HZA9 A0A3B0K1U1 W5JUF5 U4UAE4 B4JDE1 A0A1W4V686 A0A2M4BCH1 A0A2R5LMY8 A0A2M4A9M8 A0A0J9QUH0 A0A1A9X617 A0A1A9V6W4 A0A1Z5LDR3 B3NA54 B3N1A4 B4I2P7 B4NWU1 A0A1A9WDM6 T1JHK8 A0A1B0A5Y0 A0A1B0FQV4 B4LUU6 A0A1S3K2Y4 Q9VQG0 B4KLD4
A0A1B6LXV9 A0A0T6AX32 A0A1B6FRA0 A0A023F2N1 A0A0A9ZA41 A0A0K8SR47 A0A1B6DC68 A0A1B6I755 T1I4V9 D2A1S2 A0A154PB99 A0A1Y1N445 A0A0C9R8X6 K7IQL9 A0A232FHY9 A0A1W4XUS1 A0A026W543 E2ACK2 E2B5G1 A0A2A3EF25 E0VRR5 A0A0M9A2Y2 A0A151JME7 A0A0L7RE98 A0A158NXU8 Q17F50 A0A1S3DGY8 A0A1I8MGA5 A0A182GE21 A0A182H7T2 A0A182M6K7 A0A0P6ITY6 A0A1Q3F9Q7 A0A1Q3F9K5 A0A182VQ52 B0WT06 A0A151HY19 A0A182K3M2 A0A195FYK9 A0A131Y0D2 A0A0L8HPQ8 A0A182PS22 A0A2T7P5Z6 A0A0L0CFK2 A0A182VJJ5 Q5TT59 A0A182U4N2 A0A182Y7D7 A0A182KXI0 A0A0K8UQ20 A0A0A1WFH3 A0A182T4N5 A0A1Q3F9P6 A0A1Q3F9K1 A0A182Q5Y4 B4N0H1 A0A084VYU2 A0A182NLH0 A0A1I8P2Y7 A0A182FT12 A0A182RZG6 Q29KW7 B4GSP8 A0A2C9H8L6 A0A1S4GJ51 A0A182HZA9 A0A3B0K1U1 W5JUF5 U4UAE4 B4JDE1 A0A1W4V686 A0A2M4BCH1 A0A2R5LMY8 A0A2M4A9M8 A0A0J9QUH0 A0A1A9X617 A0A1A9V6W4 A0A1Z5LDR3 B3NA54 B3N1A4 B4I2P7 B4NWU1 A0A1A9WDM6 T1JHK8 A0A1B0A5Y0 A0A1B0FQV4 B4LUU6 A0A1S3K2Y4 Q9VQG0 B4KLD4
Pubmed
28756777
22118469
24845553
25474469
25401762
18362917
+ More
19820115 28004739 20075255 28648823 24508170 30249741 20798317 20566863 21347285 17510324 25315136 26483478 26999592 29652888 26108605 12364791 25244985 20966253 25830018 17994087 24438588 15632085 20920257 23761445 23537049 22936249 28528879 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
19820115 28004739 20075255 28648823 24508170 30249741 20798317 20566863 21347285 17510324 25315136 26483478 26999592 29652888 26108605 12364791 25244985 20966253 25830018 17994087 24438588 15632085 20920257 23761445 23537049 22936249 28528879 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
NWSH01000849
PCG73863.1
KZ150222
PZC72091.1
RSAL01000043
RVE50748.1
+ More
AGBW02010785 OWR47750.1 NEVH01005887 PNF38379.1 KK852758 KDR17047.1 GEBQ01011533 JAT28444.1 LJIG01022615 KRT79617.1 GECZ01017059 JAS52710.1 GBBI01003418 JAC15294.1 GBHO01002823 JAG40781.1 GBRD01009972 JAG55852.1 GEDC01024226 GEDC01014005 JAS13072.1 JAS23293.1 GECU01024969 JAS82737.1 ACPB03010532 KQ971338 EFA02119.2 KQ434864 KZC09121.1 GEZM01013329 JAV92653.1 GBYB01012799 JAG82566.1 NNAY01000191 OXU30130.1 KK107419 QOIP01000003 EZA51113.1 RLU25209.1 GL438539 EFN68837.1 GL445827 EFN89047.1 KZ288269 PBC29882.1 DS235494 EEB16071.1 KQ435765 KOX75356.1 KQ978931 KYN27455.1 KQ414613 KOC69056.1 ADTU01003566 ADTU01003567 ADTU01003568 CH477276 EAT45133.1 JXUM01056925 JXUM01056926 JXUM01056927 JXUM01056928 JXUM01056929 KQ561931 KXJ77130.1 JXUM01116950 JXUM01116951 JXUM01116952 JXUM01116953 JXUM01116954 JXUM01116955 KQ566037 KXJ70442.1 AXCM01001356 GDUN01000639 JAN95280.1 GFDL01010734 JAV24311.1 GFDL01010784 JAV24261.1 DS232079 EDS34143.1 KQ976735 KYM76136.1 KQ981204 KYN44919.1 GEFM01004465 GEGO01002042 JAP71331.1 JAR93362.1 KQ417611 KOF91139.1 PZQS01000006 PVD28826.1 JRES01000438 KNC31193.1 AAAB01008888 EAL40594.1 GDHF01023540 JAI28774.1 GBXI01016866 JAC97425.1 GFDL01010744 JAV24301.1 GFDL01010867 JAV24178.1 AXCN02000136 CH963920 EDW77584.1 ATLV01018466 KE525234 KFB43136.1 CH379061 EAL33057.2 CH479189 EDW25407.1 APCN01005522 OUUW01000006 SPP81850.1 ADMH02000418 ETN66595.1 KB632261 ERL90864.1 CH916368 EDW03311.1 GGFJ01001608 MBW50749.1 GGLE01006756 MBY10882.1 GGFK01004182 MBW37503.1 CM002910 KMY87728.1 GFJQ02001806 JAW05164.1 CH954177 EDV57517.1 CH902652 EDV33625.1 CH480820 EDW54042.1 CM000157 EDW87433.1 JH429636 CCAG010010711 CH940649 EDW64273.1 AE014134 AY095186 AAF51214.2 AAM12279.1 AGB92498.1 CH933807 EDW12815.1
AGBW02010785 OWR47750.1 NEVH01005887 PNF38379.1 KK852758 KDR17047.1 GEBQ01011533 JAT28444.1 LJIG01022615 KRT79617.1 GECZ01017059 JAS52710.1 GBBI01003418 JAC15294.1 GBHO01002823 JAG40781.1 GBRD01009972 JAG55852.1 GEDC01024226 GEDC01014005 JAS13072.1 JAS23293.1 GECU01024969 JAS82737.1 ACPB03010532 KQ971338 EFA02119.2 KQ434864 KZC09121.1 GEZM01013329 JAV92653.1 GBYB01012799 JAG82566.1 NNAY01000191 OXU30130.1 KK107419 QOIP01000003 EZA51113.1 RLU25209.1 GL438539 EFN68837.1 GL445827 EFN89047.1 KZ288269 PBC29882.1 DS235494 EEB16071.1 KQ435765 KOX75356.1 KQ978931 KYN27455.1 KQ414613 KOC69056.1 ADTU01003566 ADTU01003567 ADTU01003568 CH477276 EAT45133.1 JXUM01056925 JXUM01056926 JXUM01056927 JXUM01056928 JXUM01056929 KQ561931 KXJ77130.1 JXUM01116950 JXUM01116951 JXUM01116952 JXUM01116953 JXUM01116954 JXUM01116955 KQ566037 KXJ70442.1 AXCM01001356 GDUN01000639 JAN95280.1 GFDL01010734 JAV24311.1 GFDL01010784 JAV24261.1 DS232079 EDS34143.1 KQ976735 KYM76136.1 KQ981204 KYN44919.1 GEFM01004465 GEGO01002042 JAP71331.1 JAR93362.1 KQ417611 KOF91139.1 PZQS01000006 PVD28826.1 JRES01000438 KNC31193.1 AAAB01008888 EAL40594.1 GDHF01023540 JAI28774.1 GBXI01016866 JAC97425.1 GFDL01010744 JAV24301.1 GFDL01010867 JAV24178.1 AXCN02000136 CH963920 EDW77584.1 ATLV01018466 KE525234 KFB43136.1 CH379061 EAL33057.2 CH479189 EDW25407.1 APCN01005522 OUUW01000006 SPP81850.1 ADMH02000418 ETN66595.1 KB632261 ERL90864.1 CH916368 EDW03311.1 GGFJ01001608 MBW50749.1 GGLE01006756 MBY10882.1 GGFK01004182 MBW37503.1 CM002910 KMY87728.1 GFJQ02001806 JAW05164.1 CH954177 EDV57517.1 CH902652 EDV33625.1 CH480820 EDW54042.1 CM000157 EDW87433.1 JH429636 CCAG010010711 CH940649 EDW64273.1 AE014134 AY095186 AAF51214.2 AAM12279.1 AGB92498.1 CH933807 EDW12815.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000235965
UP000027135
UP000015103
+ More
UP000007266 UP000076502 UP000002358 UP000215335 UP000192223 UP000053097 UP000279307 UP000000311 UP000008237 UP000242457 UP000009046 UP000053105 UP000078492 UP000053825 UP000005205 UP000008820 UP000079169 UP000095301 UP000069940 UP000249989 UP000075883 UP000075920 UP000002320 UP000078540 UP000075881 UP000078541 UP000053454 UP000075885 UP000245119 UP000037069 UP000075903 UP000007062 UP000075902 UP000076408 UP000075882 UP000075901 UP000075886 UP000007798 UP000030765 UP000075884 UP000095300 UP000069272 UP000075900 UP000001819 UP000008744 UP000076407 UP000075840 UP000268350 UP000000673 UP000030742 UP000001070 UP000192221 UP000092443 UP000078200 UP000008711 UP000007801 UP000001292 UP000002282 UP000091820 UP000092445 UP000092444 UP000008792 UP000085678 UP000000803 UP000009192
UP000007266 UP000076502 UP000002358 UP000215335 UP000192223 UP000053097 UP000279307 UP000000311 UP000008237 UP000242457 UP000009046 UP000053105 UP000078492 UP000053825 UP000005205 UP000008820 UP000079169 UP000095301 UP000069940 UP000249989 UP000075883 UP000075920 UP000002320 UP000078540 UP000075881 UP000078541 UP000053454 UP000075885 UP000245119 UP000037069 UP000075903 UP000007062 UP000075902 UP000076408 UP000075882 UP000075901 UP000075886 UP000007798 UP000030765 UP000075884 UP000095300 UP000069272 UP000075900 UP000001819 UP000008744 UP000076407 UP000075840 UP000268350 UP000000673 UP000030742 UP000001070 UP000192221 UP000092443 UP000078200 UP000008711 UP000007801 UP000001292 UP000002282 UP000091820 UP000092445 UP000092444 UP000008792 UP000085678 UP000000803 UP000009192
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JQY8
A0A2W1BCF1
A0A3S2P2K2
A0A212F1Y2
A0A2J7RC24
A0A067R215
+ More
A0A1B6LXV9 A0A0T6AX32 A0A1B6FRA0 A0A023F2N1 A0A0A9ZA41 A0A0K8SR47 A0A1B6DC68 A0A1B6I755 T1I4V9 D2A1S2 A0A154PB99 A0A1Y1N445 A0A0C9R8X6 K7IQL9 A0A232FHY9 A0A1W4XUS1 A0A026W543 E2ACK2 E2B5G1 A0A2A3EF25 E0VRR5 A0A0M9A2Y2 A0A151JME7 A0A0L7RE98 A0A158NXU8 Q17F50 A0A1S3DGY8 A0A1I8MGA5 A0A182GE21 A0A182H7T2 A0A182M6K7 A0A0P6ITY6 A0A1Q3F9Q7 A0A1Q3F9K5 A0A182VQ52 B0WT06 A0A151HY19 A0A182K3M2 A0A195FYK9 A0A131Y0D2 A0A0L8HPQ8 A0A182PS22 A0A2T7P5Z6 A0A0L0CFK2 A0A182VJJ5 Q5TT59 A0A182U4N2 A0A182Y7D7 A0A182KXI0 A0A0K8UQ20 A0A0A1WFH3 A0A182T4N5 A0A1Q3F9P6 A0A1Q3F9K1 A0A182Q5Y4 B4N0H1 A0A084VYU2 A0A182NLH0 A0A1I8P2Y7 A0A182FT12 A0A182RZG6 Q29KW7 B4GSP8 A0A2C9H8L6 A0A1S4GJ51 A0A182HZA9 A0A3B0K1U1 W5JUF5 U4UAE4 B4JDE1 A0A1W4V686 A0A2M4BCH1 A0A2R5LMY8 A0A2M4A9M8 A0A0J9QUH0 A0A1A9X617 A0A1A9V6W4 A0A1Z5LDR3 B3NA54 B3N1A4 B4I2P7 B4NWU1 A0A1A9WDM6 T1JHK8 A0A1B0A5Y0 A0A1B0FQV4 B4LUU6 A0A1S3K2Y4 Q9VQG0 B4KLD4
A0A1B6LXV9 A0A0T6AX32 A0A1B6FRA0 A0A023F2N1 A0A0A9ZA41 A0A0K8SR47 A0A1B6DC68 A0A1B6I755 T1I4V9 D2A1S2 A0A154PB99 A0A1Y1N445 A0A0C9R8X6 K7IQL9 A0A232FHY9 A0A1W4XUS1 A0A026W543 E2ACK2 E2B5G1 A0A2A3EF25 E0VRR5 A0A0M9A2Y2 A0A151JME7 A0A0L7RE98 A0A158NXU8 Q17F50 A0A1S3DGY8 A0A1I8MGA5 A0A182GE21 A0A182H7T2 A0A182M6K7 A0A0P6ITY6 A0A1Q3F9Q7 A0A1Q3F9K5 A0A182VQ52 B0WT06 A0A151HY19 A0A182K3M2 A0A195FYK9 A0A131Y0D2 A0A0L8HPQ8 A0A182PS22 A0A2T7P5Z6 A0A0L0CFK2 A0A182VJJ5 Q5TT59 A0A182U4N2 A0A182Y7D7 A0A182KXI0 A0A0K8UQ20 A0A0A1WFH3 A0A182T4N5 A0A1Q3F9P6 A0A1Q3F9K1 A0A182Q5Y4 B4N0H1 A0A084VYU2 A0A182NLH0 A0A1I8P2Y7 A0A182FT12 A0A182RZG6 Q29KW7 B4GSP8 A0A2C9H8L6 A0A1S4GJ51 A0A182HZA9 A0A3B0K1U1 W5JUF5 U4UAE4 B4JDE1 A0A1W4V686 A0A2M4BCH1 A0A2R5LMY8 A0A2M4A9M8 A0A0J9QUH0 A0A1A9X617 A0A1A9V6W4 A0A1Z5LDR3 B3NA54 B3N1A4 B4I2P7 B4NWU1 A0A1A9WDM6 T1JHK8 A0A1B0A5Y0 A0A1B0FQV4 B4LUU6 A0A1S3K2Y4 Q9VQG0 B4KLD4
PDB
4C0P
E-value=2.96113e-154,
Score=1402
Ontologies
GO
Topology
Subcellular location
Nucleus
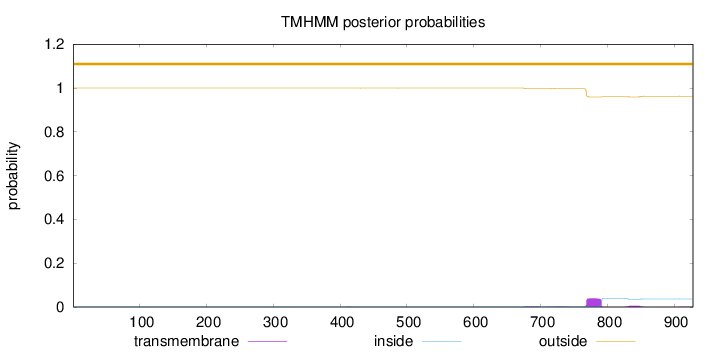
Length:
928
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.09183
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00030
outside
1 - 928
Population Genetic Test Statistics
Pi
258.226268
Theta
211.914839
Tajima's D
0.846024
CLR
0.263454
CSRT
0.612619369031548
Interpretation
Uncertain
