Pre Gene Modal
BGIBMGA010495
Annotation
PREDICTED:_chromatin_assembly_factor_1_subunit_B_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.236
Sequence
CDS
ATGAAGTTTGCGATACCAGAAATATCCTGGCATAATCGTGATCCAGTTCTAAGTGTAGATTTTCAGCCCAAAGCGGAACCGAATGACCCGTTACGACTCGCCAGCGGTGGAACAGATTCACACGTTTTGATCTGGTATTTATCAACCACGGAGACGGGTTCAGTTCATCTTGAAGTAGCAGCTGATTTAACAAGGCATCAGAAAGCTGTGAATGTAGTGAGATGGTCACCTAATGGACAGTTATTGGCTTCTGGGGATGATGAGTCCATCATATTCATTTGGAAGCAGAAACTGGAAGAACTTTCTCCGGCAACTGAGGGTGAGGAGCAATATAAAGAAACTTGGGTTGTTTATAAGACTCTTCGAGGGCACATGGAAGATGTCCTTGACATATCCTGGAGTATGGACGGCTTACAGTTGGCTTCTGGCTCAGTAGATAACAAATTAATTGTTTGGGACATACACAGAGCTCGTTACACTACTATACTTAGTGACCACAAAGGGTTTGTGCAAGGTGTCTGTTGGGATCCTAAAGGACAGTACATAACATCTACGAGTACAGACAGAGTGTTTCGAGCATTTGATGTATCTACCAAGAAGGTTGTATCAAGAAGCAGCAAAGCATCTCTCCCGTTCCCCACTGACCATCCTCTCCACGGCTCCAGACAACGGCTATACCATGATGACACACTCCAAACATATTACCGACGACTGCAGTTCAGCCCGGACGGCCTATACATTGCAGTTCCAGCAGGGAGAGTGGAGCCAGAGGGAAAACCTGATGTAAAACCAATCAATGCCGTGTATATCTATACGAGATATTGTTTGAAAATCCCAGTGTGTGTGGTACCATGCGGCGAGCCAGCCCTGGTGACCCGCTGGTCGCCGATAGGGTTCCGCCCCCGCGCCGCCGGGCCCGCGCCTCGCCTGCCGTCCCCGAGCGGAGCCCGGCACGTGCTGGCCGTGGGGACCAGGCGCTCCGTGTTGGTGTACGACACACAACAGAAGACTCCCATAGCCATAGTTGCCAATGTACATTACACTAGATTAACAGATCTAACATGGTCCAACAATGGTCGTATACTGGTGGCCTCCAGCACTGACGGCTTCTGCTCCATTATCACATTCAACAATGAGGAACTCGGCGAGCCACTTCCTCTAGAACCAAACACTGAGAATAAACCAGAACCAATGGAAACGCAAGACGAAAAGCCTGCAGCTCAAACAGAAGCGCCGAAACCGACAAAAATAGATTCGTTCATAAAATTCAAAGCGCCAGAAAAATCGCCCAAAAATTTAAAAACACAAACAGAAATCAAAACGCCCGTCAAAATAGACTTTTTAGAGGAAGTCGCCAACCCCTCGTGGTCCGATAACAGTAACAACGAAATTATAAAACATCGAGCGAGCGAACCGATGGACTCCGACGTGACGGTCATAGAAGACAGCGGCGATATAAAACTGGTTTACGAGGAAACTGAAGGCGTCAAGTCGCCGCCTAAAACGCAACAAGCCAACAAACTGGAAATTAAAAGTAAACAAAACACTTCGCCCAAAGACAATCAGTTCTTTAGACAGGCCAAGGTGACAGATGTCAAACAGCCAATAGCGGCAACAGTGTCCAGTCCTAAAGCGCCTCGGAGGGTCAGCTTCGTAACTTTGTCAAGTCCTAAAAATGCTAAAAAGAAATAG
Protein
MKFAIPEISWHNRDPVLSVDFQPKAEPNDPLRLASGGTDSHVLIWYLSTTETGSVHLEVAADLTRHQKAVNVVRWSPNGQLLASGDDESIIFIWKQKLEELSPATEGEEQYKETWVVYKTLRGHMEDVLDISWSMDGLQLASGSVDNKLIVWDIHRARYTTILSDHKGFVQGVCWDPKGQYITSTSTDRVFRAFDVSTKKVVSRSSKASLPFPTDHPLHGSRQRLYHDDTLQTYYRRLQFSPDGLYIAVPAGRVEPEGKPDVKPINAVYIYTRYCLKIPVCVVPCGEPALVTRWSPIGFRPRAAGPAPRLPSPSGARHVLAVGTRRSVLVYDTQQKTPIAIVANVHYTRLTDLTWSNNGRILVASSTDGFCSIITFNNEELGEPLPLEPNTENKPEPMETQDEKPAAQTEAPKPTKIDSFIKFKAPEKSPKNLKTQTEIKTPVKIDFLEEVANPSWSDNSNNEIIKHRASEPMDSDVTVIEDSGDIKLVYEETEGVKSPPKTQQANKLEIKSKQNTSPKDNQFFRQAKVTDVKQPIAATVSSPKAPRRVSFVTLSSPKNAKKK
Summary
Uniprot
Pubmed
EMBL
BABH01005121
KZ150222
PZC72090.1
NWSH01000849
PCG73862.1
ODYU01008383
+ More
SOQ51930.1 AGBW02010785 OWR47751.1 KQ461194 KPJ06834.1 KQ458671 KPJ05522.1 JTDY01002245 KOB71813.1 UFQT01001176 SSX29341.1 UFQT01000530 SSX25025.1 AJVK01029976 KZ288292 PBC29186.1 KQ435944 KOX68197.1 KQ414596 KOC70042.1 CH963719 EDW71991.1 KQ971312 EEZ98093.1 KQ434864 KZC09000.1 NNAY01000180 OXU30243.1 KK107151 QOIP01000003 EZA57248.1 RLU24388.1 ADTU01003493 ADTU01003494 KQ976462 KYM84657.1 LBMM01003848 KMQ93058.1 KQ981015 KYN10274.1 GL888216 EGI64810.1 KQ981204 KYN44973.1 GL452008 EFN78079.1
SOQ51930.1 AGBW02010785 OWR47751.1 KQ461194 KPJ06834.1 KQ458671 KPJ05522.1 JTDY01002245 KOB71813.1 UFQT01001176 SSX29341.1 UFQT01000530 SSX25025.1 AJVK01029976 KZ288292 PBC29186.1 KQ435944 KOX68197.1 KQ414596 KOC70042.1 CH963719 EDW71991.1 KQ971312 EEZ98093.1 KQ434864 KZC09000.1 NNAY01000180 OXU30243.1 KK107151 QOIP01000003 EZA57248.1 RLU24388.1 ADTU01003493 ADTU01003494 KQ976462 KYM84657.1 LBMM01003848 KMQ93058.1 KQ981015 KYN10274.1 GL888216 EGI64810.1 KQ981204 KYN44973.1 GL452008 EFN78079.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000092462 UP000242457 UP000053105 UP000005203 UP000053825 UP000007798 UP000007266 UP000076502 UP000215335 UP000002358 UP000053097 UP000279307 UP000005205 UP000078540 UP000036403 UP000078492 UP000007755 UP000078541 UP000008237
UP000092462 UP000242457 UP000053105 UP000005203 UP000053825 UP000007798 UP000007266 UP000076502 UP000215335 UP000002358 UP000053097 UP000279307 UP000005205 UP000078540 UP000036403 UP000078492 UP000007755 UP000078541 UP000008237
PRIDE
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
PDB
4CY5
E-value=1.13333e-11,
Score=170
Ontologies
GO
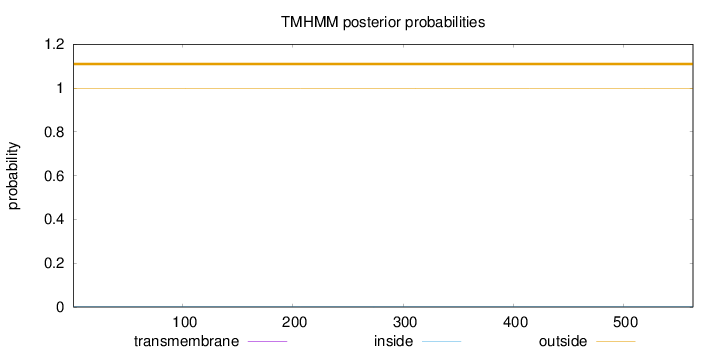
Topology
Length:
563
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00181
Exp number, first 60 AAs:
0.00057
Total prob of N-in:
0.00085
outside
1 - 563
Population Genetic Test Statistics
Pi
337.163423
Theta
234.164605
Tajima's D
1.385317
CLR
0.000379
CSRT
0.75821208939553
Interpretation
Uncertain
