Pre Gene Modal
BGIBMGA010488
Annotation
PREDICTED:_pre-mRNA-splicing_factor_SPF27_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.258
Sequence
CDS
ATGGCAGGAGAAGTTGTCGTAGATGCATTACCATATATAGACCAAGGATATGACGATCCTGGAGTAAGAGAAGCTGCATTGGCCATGGTGGAAGAAGAGTGCAGACGCTATAGACCCACAAAGAACTATCTTGAAAATGCAGGACCAGAACCAAGTTCTGCATTTGAAACTCCTGCTTTACAACGAGAGATGGAAAGAGTGCAACAGAGATTGCCGATGGAACCATTGTCTATGAAGAGGTATGAACTTCCACCCCCTCCAGTCGGAAGAATGAGTGAACCAAGTGCCTGGGCCGAGTCAGTTGACAACTCTCATGCACAGCTGTCACACCAAGCTATCAGAGTTCTCAATTTAGAGTTGCAGTTAGAGTACGGTTCAGAAGCATGGAGATCATACCTCAATACACTTCAAGCATTAGTCACAAGAGCTCAAAACGAACACAACCAGTTACGTAAACAAATAGGCTCTGTGAACTGGGAACGTAAACGCGTACAAACAGCAAGCGGTGCGAGGTTGCGGGCATTGAGGCGACGGTGGGCGCAACTTGTCTCGGACAATTATAGACTTGAAAGAGCCCTACTTCACTTACAACAGGCAAAGGGAGAAGTACCAGTAGAAGAAGGAGAAGAAGCCGATTTGGACATTGTAAAGTTACCTGAAAAACTCAATTCGGAAACAGAGAAGGAAGTGCAAAAGAAAATTGAAGATATGTTTGCGGATTAA
Protein
MAGEVVVDALPYIDQGYDDPGVREAALAMVEEECRRYRPTKNYLENAGPEPSSAFETPALQREMERVQQRLPMEPLSMKRYELPPPPVGRMSEPSAWAESVDNSHAQLSHQAIRVLNLELQLEYGSEAWRSYLNTLQALVTRAQNEHNQLRKQIGSVNWERKRVQTASGARLRALRRRWAQLVSDNYRLERALLHLQQAKGEVPVEEGEEADLDIVKLPEKLNSETEKEVQKKIEDMFAD
Summary
Uniprot
H9JLT6
A0A2W1BFT6
A0A2H1WFY7
A0A2A4JPW5
A0A194QTF8
A0A194QJF8
+ More
A0A212F208 S4Q0A5 A0A076FHL3 A0A0M8ZYS3 A0A0L7QYH9 A0A154PLY9 A0A0L0CH84 Q16U78 A0A2A3EUZ9 A0A0B4J2N9 A0A067RGW9 B0WA23 A0A182JZB7 E2C704 A0A151JVH2 A0A2M4AYM5 A0A1Q3F3K4 A0A182SYY2 A0A151J691 A0A182NG96 A0A182XLZ7 A0A195BB77 A0A158NIM3 A0A182VCL5 A0A2J7QLQ8 A0A182JHP5 A0A182XVE8 A0A182UFJ1 A0A182LFN1 Q7QBT8 A0A182HTH1 A0A182PEL7 A0A1I8PUC9 A0A182FQ07 A0A151ILY0 A0A2M4AXQ0 W5JER8 A0A0J7KLM0 A0A1B6FQK1 A0A2M4BYV1 A0A182M245 U5ETJ1 A0A2M3ZCQ7 A0A2M4BYU4 A0A182W2M5 A0A1B6KAV1 A0A026WL09 A0A182Q497 E2A9S4 A0A151WQ78 A0A182R759 A0A1B6DPE7 A0A1S3D4Y6 B3M016 B4JEQ7 A0A1I8M2M0 F4WDL9 T1HTR9 A0A0N7Z8I8 A0A224XXK6 A0A023F585 A0A069DQ66 A0A0V0G8N5 A0A0B6XZ98 D6WKN2 A0A2H8TFM7 A0A232EE42 A0A161MQ43 J9K9X3 A0A1Y1NCX3 A0A034VG38 E0VPG5 N6U094 J3JZ00 T1EB99 A0A0M4EGN4 C4WW46 A0A0A9X2Y5 A0A1A9XG49 A0A1B0BEH2 D3TRT2 A0A2L2YG07 A0A1A9Z9K7 A0A1W4X679 A0A1A9UJQ4 A0A0K8VA35 A0A1L8DHP1 A0A0L8IB39 B3P5W1 B4G2B6 A0A1L8DHH4 B4PR85 A0A1A9WY30 V4AVU3
A0A212F208 S4Q0A5 A0A076FHL3 A0A0M8ZYS3 A0A0L7QYH9 A0A154PLY9 A0A0L0CH84 Q16U78 A0A2A3EUZ9 A0A0B4J2N9 A0A067RGW9 B0WA23 A0A182JZB7 E2C704 A0A151JVH2 A0A2M4AYM5 A0A1Q3F3K4 A0A182SYY2 A0A151J691 A0A182NG96 A0A182XLZ7 A0A195BB77 A0A158NIM3 A0A182VCL5 A0A2J7QLQ8 A0A182JHP5 A0A182XVE8 A0A182UFJ1 A0A182LFN1 Q7QBT8 A0A182HTH1 A0A182PEL7 A0A1I8PUC9 A0A182FQ07 A0A151ILY0 A0A2M4AXQ0 W5JER8 A0A0J7KLM0 A0A1B6FQK1 A0A2M4BYV1 A0A182M245 U5ETJ1 A0A2M3ZCQ7 A0A2M4BYU4 A0A182W2M5 A0A1B6KAV1 A0A026WL09 A0A182Q497 E2A9S4 A0A151WQ78 A0A182R759 A0A1B6DPE7 A0A1S3D4Y6 B3M016 B4JEQ7 A0A1I8M2M0 F4WDL9 T1HTR9 A0A0N7Z8I8 A0A224XXK6 A0A023F585 A0A069DQ66 A0A0V0G8N5 A0A0B6XZ98 D6WKN2 A0A2H8TFM7 A0A232EE42 A0A161MQ43 J9K9X3 A0A1Y1NCX3 A0A034VG38 E0VPG5 N6U094 J3JZ00 T1EB99 A0A0M4EGN4 C4WW46 A0A0A9X2Y5 A0A1A9XG49 A0A1B0BEH2 D3TRT2 A0A2L2YG07 A0A1A9Z9K7 A0A1W4X679 A0A1A9UJQ4 A0A0K8VA35 A0A1L8DHP1 A0A0L8IB39 B3P5W1 B4G2B6 A0A1L8DHH4 B4PR85 A0A1A9WY30 V4AVU3
Pubmed
19121390
28756777
26354079
22118469
23622113
26108605
+ More
17510324 24845553 20798317 21347285 25244985 20966253 12364791 14747013 17210077 20920257 23761445 24508170 30249741 17994087 25315136 21719571 27129103 25474469 26334808 18362917 19820115 28648823 28004739 25348373 20566863 23537049 22516182 25401762 20353571 26561354 17550304 23254933
17510324 24845553 20798317 21347285 25244985 20966253 12364791 14747013 17210077 20920257 23761445 24508170 30249741 17994087 25315136 21719571 27129103 25474469 26334808 18362917 19820115 28648823 28004739 25348373 20566863 23537049 22516182 25401762 20353571 26561354 17550304 23254933
EMBL
BABH01005119
BABH01005120
KZ150222
PZC72087.1
ODYU01008383
SOQ51927.1
+ More
NWSH01000849 PCG73859.1 KQ461194 KPJ06831.1 KQ458671 KPJ05524.1 AGBW02010785 OWR47754.1 GAIX01000128 JAA92432.1 KF803644 AII16886.1 KQ435821 KOX72329.1 KQ414688 KOC63649.1 KQ434948 KZC12464.1 JRES01000409 KNC31601.1 CH477631 EAT38055.1 KZ288185 PBC34891.1 KK852478 KDR23022.1 DS231868 EDS40756.1 GL453255 EFN76276.1 KQ981702 KYN37454.1 GGFK01012377 MBW45698.1 GFDL01012922 JAV22123.1 KQ979869 KYN18737.1 KQ976529 KYM81806.1 ADTU01017000 NEVH01013241 PNF29517.1 AAAB01008859 EAA07483.3 APCN01000528 KQ977085 KYN05890.1 GGFK01012229 MBW45550.1 ADMH02001428 ETN62566.1 LBMM01005671 KMQ91303.1 GECZ01017335 JAS52434.1 GGFJ01009098 MBW58239.1 AXCM01000235 GANO01002759 JAB57112.1 GGFM01005528 MBW26279.1 GGFJ01009099 MBW58240.1 GEBQ01031406 JAT08571.1 KK107159 QOIP01000001 EZA56742.1 RLU27283.1 AXCN02001627 GL437918 EFN69876.1 KQ982850 KYQ49961.1 GEDC01009769 JAS27529.1 CH902617 EDV44206.1 CH916369 EDV93188.1 GL888087 EGI67799.1 ACPB03011663 GDKW01003227 JAI53368.1 GFTR01003627 JAW12799.1 GBBI01002459 JAC16253.1 GBGD01002884 JAC86005.1 GECL01001843 JAP04281.1 HACG01001986 CEK48851.1 KQ971342 EFA03021.1 GFXV01000717 MBW12522.1 NNAY01005655 OXU16598.1 GEMB01002650 JAS00539.1 ABLF02003796 ABLF02009129 GEZM01006507 JAV95743.1 GAKP01018237 JAC40715.1 DS235366 EEB15271.1 APGK01054266 KB741248 KB632305 ENN71957.1 ERL91844.1 BT128481 AEE63438.1 GAMD01000068 JAB01523.1 CP012526 ALC47566.1 AK341845 BAH72116.1 GBHO01032129 JAG11475.1 JXJN01012899 CCAG010014727 EZ424134 ADD20410.1 IAAA01013251 LAA06165.1 GDHF01033524 GDHF01023230 GDHF01016500 JAI18790.1 JAI29084.1 JAI35814.1 GFDF01008177 JAV05907.1 KQ416196 KOF98235.1 CH954182 EDV53361.1 CH479179 EDW23961.1 GFDF01008176 JAV05908.1 CM000160 EDW98449.1 KB200505 ESP01498.1
NWSH01000849 PCG73859.1 KQ461194 KPJ06831.1 KQ458671 KPJ05524.1 AGBW02010785 OWR47754.1 GAIX01000128 JAA92432.1 KF803644 AII16886.1 KQ435821 KOX72329.1 KQ414688 KOC63649.1 KQ434948 KZC12464.1 JRES01000409 KNC31601.1 CH477631 EAT38055.1 KZ288185 PBC34891.1 KK852478 KDR23022.1 DS231868 EDS40756.1 GL453255 EFN76276.1 KQ981702 KYN37454.1 GGFK01012377 MBW45698.1 GFDL01012922 JAV22123.1 KQ979869 KYN18737.1 KQ976529 KYM81806.1 ADTU01017000 NEVH01013241 PNF29517.1 AAAB01008859 EAA07483.3 APCN01000528 KQ977085 KYN05890.1 GGFK01012229 MBW45550.1 ADMH02001428 ETN62566.1 LBMM01005671 KMQ91303.1 GECZ01017335 JAS52434.1 GGFJ01009098 MBW58239.1 AXCM01000235 GANO01002759 JAB57112.1 GGFM01005528 MBW26279.1 GGFJ01009099 MBW58240.1 GEBQ01031406 JAT08571.1 KK107159 QOIP01000001 EZA56742.1 RLU27283.1 AXCN02001627 GL437918 EFN69876.1 KQ982850 KYQ49961.1 GEDC01009769 JAS27529.1 CH902617 EDV44206.1 CH916369 EDV93188.1 GL888087 EGI67799.1 ACPB03011663 GDKW01003227 JAI53368.1 GFTR01003627 JAW12799.1 GBBI01002459 JAC16253.1 GBGD01002884 JAC86005.1 GECL01001843 JAP04281.1 HACG01001986 CEK48851.1 KQ971342 EFA03021.1 GFXV01000717 MBW12522.1 NNAY01005655 OXU16598.1 GEMB01002650 JAS00539.1 ABLF02003796 ABLF02009129 GEZM01006507 JAV95743.1 GAKP01018237 JAC40715.1 DS235366 EEB15271.1 APGK01054266 KB741248 KB632305 ENN71957.1 ERL91844.1 BT128481 AEE63438.1 GAMD01000068 JAB01523.1 CP012526 ALC47566.1 AK341845 BAH72116.1 GBHO01032129 JAG11475.1 JXJN01012899 CCAG010014727 EZ424134 ADD20410.1 IAAA01013251 LAA06165.1 GDHF01033524 GDHF01023230 GDHF01016500 JAI18790.1 JAI29084.1 JAI35814.1 GFDF01008177 JAV05907.1 KQ416196 KOF98235.1 CH954182 EDV53361.1 CH479179 EDW23961.1 GFDF01008176 JAV05908.1 CM000160 EDW98449.1 KB200505 ESP01498.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000053105
+ More
UP000053825 UP000076502 UP000037069 UP000008820 UP000242457 UP000005203 UP000027135 UP000002320 UP000075881 UP000008237 UP000078541 UP000075901 UP000078492 UP000075884 UP000076407 UP000078540 UP000005205 UP000075903 UP000235965 UP000075880 UP000076408 UP000075902 UP000075882 UP000007062 UP000075840 UP000075885 UP000095300 UP000069272 UP000078542 UP000000673 UP000036403 UP000075883 UP000075920 UP000053097 UP000279307 UP000075886 UP000000311 UP000075809 UP000075900 UP000079169 UP000007801 UP000001070 UP000095301 UP000007755 UP000015103 UP000007266 UP000215335 UP000007819 UP000009046 UP000019118 UP000030742 UP000092553 UP000092443 UP000092460 UP000092444 UP000092445 UP000192223 UP000078200 UP000053454 UP000008711 UP000008744 UP000002282 UP000091820 UP000030746
UP000053825 UP000076502 UP000037069 UP000008820 UP000242457 UP000005203 UP000027135 UP000002320 UP000075881 UP000008237 UP000078541 UP000075901 UP000078492 UP000075884 UP000076407 UP000078540 UP000005205 UP000075903 UP000235965 UP000075880 UP000076408 UP000075902 UP000075882 UP000007062 UP000075840 UP000075885 UP000095300 UP000069272 UP000078542 UP000000673 UP000036403 UP000075883 UP000075920 UP000053097 UP000279307 UP000075886 UP000000311 UP000075809 UP000075900 UP000079169 UP000007801 UP000001070 UP000095301 UP000007755 UP000015103 UP000007266 UP000215335 UP000007819 UP000009046 UP000019118 UP000030742 UP000092553 UP000092443 UP000092460 UP000092444 UP000092445 UP000192223 UP000078200 UP000053454 UP000008711 UP000008744 UP000002282 UP000091820 UP000030746
PRIDE
Pfam
PF05700 BCAS2
Interpro
IPR008409
SPF27
ProteinModelPortal
H9JLT6
A0A2W1BFT6
A0A2H1WFY7
A0A2A4JPW5
A0A194QTF8
A0A194QJF8
+ More
A0A212F208 S4Q0A5 A0A076FHL3 A0A0M8ZYS3 A0A0L7QYH9 A0A154PLY9 A0A0L0CH84 Q16U78 A0A2A3EUZ9 A0A0B4J2N9 A0A067RGW9 B0WA23 A0A182JZB7 E2C704 A0A151JVH2 A0A2M4AYM5 A0A1Q3F3K4 A0A182SYY2 A0A151J691 A0A182NG96 A0A182XLZ7 A0A195BB77 A0A158NIM3 A0A182VCL5 A0A2J7QLQ8 A0A182JHP5 A0A182XVE8 A0A182UFJ1 A0A182LFN1 Q7QBT8 A0A182HTH1 A0A182PEL7 A0A1I8PUC9 A0A182FQ07 A0A151ILY0 A0A2M4AXQ0 W5JER8 A0A0J7KLM0 A0A1B6FQK1 A0A2M4BYV1 A0A182M245 U5ETJ1 A0A2M3ZCQ7 A0A2M4BYU4 A0A182W2M5 A0A1B6KAV1 A0A026WL09 A0A182Q497 E2A9S4 A0A151WQ78 A0A182R759 A0A1B6DPE7 A0A1S3D4Y6 B3M016 B4JEQ7 A0A1I8M2M0 F4WDL9 T1HTR9 A0A0N7Z8I8 A0A224XXK6 A0A023F585 A0A069DQ66 A0A0V0G8N5 A0A0B6XZ98 D6WKN2 A0A2H8TFM7 A0A232EE42 A0A161MQ43 J9K9X3 A0A1Y1NCX3 A0A034VG38 E0VPG5 N6U094 J3JZ00 T1EB99 A0A0M4EGN4 C4WW46 A0A0A9X2Y5 A0A1A9XG49 A0A1B0BEH2 D3TRT2 A0A2L2YG07 A0A1A9Z9K7 A0A1W4X679 A0A1A9UJQ4 A0A0K8VA35 A0A1L8DHP1 A0A0L8IB39 B3P5W1 B4G2B6 A0A1L8DHH4 B4PR85 A0A1A9WY30 V4AVU3
A0A212F208 S4Q0A5 A0A076FHL3 A0A0M8ZYS3 A0A0L7QYH9 A0A154PLY9 A0A0L0CH84 Q16U78 A0A2A3EUZ9 A0A0B4J2N9 A0A067RGW9 B0WA23 A0A182JZB7 E2C704 A0A151JVH2 A0A2M4AYM5 A0A1Q3F3K4 A0A182SYY2 A0A151J691 A0A182NG96 A0A182XLZ7 A0A195BB77 A0A158NIM3 A0A182VCL5 A0A2J7QLQ8 A0A182JHP5 A0A182XVE8 A0A182UFJ1 A0A182LFN1 Q7QBT8 A0A182HTH1 A0A182PEL7 A0A1I8PUC9 A0A182FQ07 A0A151ILY0 A0A2M4AXQ0 W5JER8 A0A0J7KLM0 A0A1B6FQK1 A0A2M4BYV1 A0A182M245 U5ETJ1 A0A2M3ZCQ7 A0A2M4BYU4 A0A182W2M5 A0A1B6KAV1 A0A026WL09 A0A182Q497 E2A9S4 A0A151WQ78 A0A182R759 A0A1B6DPE7 A0A1S3D4Y6 B3M016 B4JEQ7 A0A1I8M2M0 F4WDL9 T1HTR9 A0A0N7Z8I8 A0A224XXK6 A0A023F585 A0A069DQ66 A0A0V0G8N5 A0A0B6XZ98 D6WKN2 A0A2H8TFM7 A0A232EE42 A0A161MQ43 J9K9X3 A0A1Y1NCX3 A0A034VG38 E0VPG5 N6U094 J3JZ00 T1EB99 A0A0M4EGN4 C4WW46 A0A0A9X2Y5 A0A1A9XG49 A0A1B0BEH2 D3TRT2 A0A2L2YG07 A0A1A9Z9K7 A0A1W4X679 A0A1A9UJQ4 A0A0K8VA35 A0A1L8DHP1 A0A0L8IB39 B3P5W1 B4G2B6 A0A1L8DHH4 B4PR85 A0A1A9WY30 V4AVU3
PDB
6QDV
E-value=3.41698e-45,
Score=455
Ontologies
GO
PANTHER
Topology
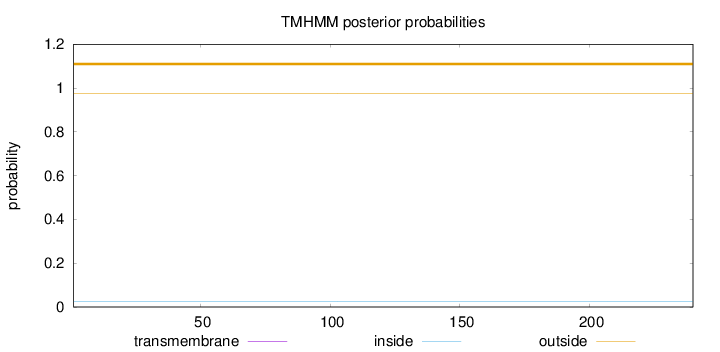
Length:
240
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02414
outside
1 - 240
Population Genetic Test Statistics
Pi
208.970449
Theta
172.35255
Tajima's D
0.621286
CLR
0.168379
CSRT
0.547972601369932
Interpretation
Uncertain
