Gene
KWMTBOMO06956
Annotation
PREDICTED:_serine_protease_snake-like_[Papilio_polytes]
Location in the cell
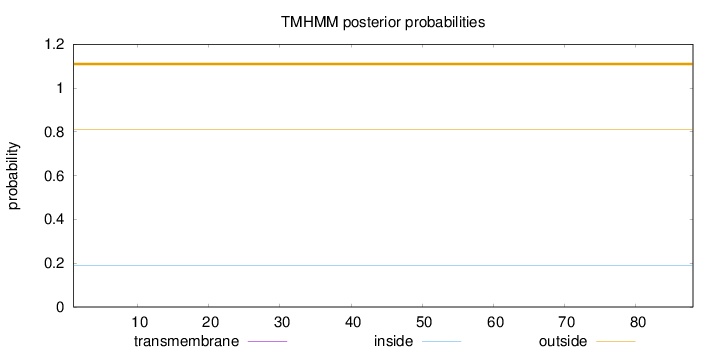
Cytoplasmic Reliability : 1.105 Nuclear Reliability : 1.645
Sequence
CDS
ATGGCACGAGTGAGTTTGATACAGGAGCCGTTTGATAACGTTGAAGTGAGCGAGTTCACCGCCGAGGAGTGCTTGACGCACTACCCTCCGCACCGCCACCTGCTCGACGGGTTCGACAACAGCACGCAGTTGTGCTACGGAGACCGCGACAAGACGCGCGACACTTGCCAGGGTGACAGCGGCGGTCCCTTGATCTTTGAAGAGGGTGAGGTGCGTTGTACGAGAACAATATTCGGCGTGATGTCATCGGGCGTGGAGTCGATGTAG
Protein
MARVSLIQEPFDNVEVSEFTAEECLTHYPPHRHLLDGFDNSTQLCYGDRDKTRDTCQGDSGGPLIFEEGEVRCTRTIFGVMSSGVESM
Summary
Similarity
Belongs to the peptidase S1 family.
Belongs to the PI3/PI4-kinase family.
Belongs to the PI3/PI4-kinase family.
Uniprot
H9JLA2
H9JLA5
H9JY18
A0A2W1B9D9
A0A2A4J4P7
A0A2A4JLI0
+ More
A0A2W1BCP3 S4NXP2 A0A2W1BHZ8 A0A2A4J4I5 A0A3S2NPZ4 A0A0N1IHL2 A0A2H1V6L6 A0A2H1VFJ7 A0A2W1BD72 Q5MPB3 A0A194QB71 A0A2A4IWJ7 A0A2H1VJG7 A0A2A4IY52 A0A2W1BC76 U5ESE3 A0A194Q0M3 A0A2H1VSY3 A0A2A4K113 A0A346RAE0 A0A0N1PHF7 A0A2W1B4D5 A0A2W1B9E2 A0A2A4K9I3 Q5MPB6 A0A0N0PDL6 A0A2A4J7N6 A0A194Q9Z8 A0A0L7KYR5 A0A2W1BAK2 H9JJA9 A0A2W1BGC1 A0A2W1B0L2 F5BZV1 A0A2W1B4D4 A0A2H1VTL8 A0A2W1B6N2 A0A2A4K2L1 A0A2W1B2E4 A0A2A4K714 Q5MPB2 A0A2H1VRC0 A0A2H1WH10 A0A2A4JMC3 A0A2W1BB28 A0A2H1WPC1 A0A2H1VFF0 A0A2A4K236 A0A2H1VS51 A0A182SUQ0 A0A2H1VA93 A0A182XUQ8 A0A182IIN2 Q5TNC5 A0A182KNG1 A0A2W1BFG9 A0A182THH3 A0A2H1W424 A0A182VNZ6 A0A2A4KAK6 A0A2W1AYZ6 A0A0L7L3B8 A0A2A4J4C8 I4DL86 A0A182S507 A0A084VYF5 A0A182Y982 A0A182JES5 A0A182Q673 A0A2W1BRV9 B0XLJ6 A0A182MQJ3 A0A2A4K9N2 A0A182JZN4 A0A336LWU0 A0A194Q9Q9 A0A182NZ25 Q16NE9 A0A2A4KAM6 A0A2M4BYH7 B0W1A9 W5JRY9 A0A182WR24 A0A182FZY7 A0A2W1B2N2 A0A1Q3FNX1 A0A2W1BDH3 A0A2W1B5J0 A0A2W1BDD3 A0A2H1VA97 A0A1B0CS98
A0A2W1BCP3 S4NXP2 A0A2W1BHZ8 A0A2A4J4I5 A0A3S2NPZ4 A0A0N1IHL2 A0A2H1V6L6 A0A2H1VFJ7 A0A2W1BD72 Q5MPB3 A0A194QB71 A0A2A4IWJ7 A0A2H1VJG7 A0A2A4IY52 A0A2W1BC76 U5ESE3 A0A194Q0M3 A0A2H1VSY3 A0A2A4K113 A0A346RAE0 A0A0N1PHF7 A0A2W1B4D5 A0A2W1B9E2 A0A2A4K9I3 Q5MPB6 A0A0N0PDL6 A0A2A4J7N6 A0A194Q9Z8 A0A0L7KYR5 A0A2W1BAK2 H9JJA9 A0A2W1BGC1 A0A2W1B0L2 F5BZV1 A0A2W1B4D4 A0A2H1VTL8 A0A2W1B6N2 A0A2A4K2L1 A0A2W1B2E4 A0A2A4K714 Q5MPB2 A0A2H1VRC0 A0A2H1WH10 A0A2A4JMC3 A0A2W1BB28 A0A2H1WPC1 A0A2H1VFF0 A0A2A4K236 A0A2H1VS51 A0A182SUQ0 A0A2H1VA93 A0A182XUQ8 A0A182IIN2 Q5TNC5 A0A182KNG1 A0A2W1BFG9 A0A182THH3 A0A2H1W424 A0A182VNZ6 A0A2A4KAK6 A0A2W1AYZ6 A0A0L7L3B8 A0A2A4J4C8 I4DL86 A0A182S507 A0A084VYF5 A0A182Y982 A0A182JES5 A0A182Q673 A0A2W1BRV9 B0XLJ6 A0A182MQJ3 A0A2A4K9N2 A0A182JZN4 A0A336LWU0 A0A194Q9Q9 A0A182NZ25 Q16NE9 A0A2A4KAM6 A0A2M4BYH7 B0W1A9 W5JRY9 A0A182WR24 A0A182FZY7 A0A2W1B2N2 A0A1Q3FNX1 A0A2W1BDH3 A0A2W1B5J0 A0A2W1BDD3 A0A2H1VA97 A0A1B0CS98
Pubmed
EMBL
BABH01038773
BABH01038774
BABH01038780
BABH01041620
BABH01041621
BABH01041622
+ More
BABH01041623 BABH01041624 KZ150484 PZC70714.1 NWSH01003268 PCG66648.1 NWSH01001045 PCG72935.1 KZ150278 PZC71645.1 GAIX01012077 JAA80483.1 KZ150344 PZC71293.1 PCG66646.1 RSAL01000491 RVE41548.1 KQ460127 KPJ17576.1 ODYU01000940 SOQ36457.1 ODYU01002282 SOQ39556.1 PZC71287.1 AY672797 AAV91019.1 KQ459249 KPJ02240.1 NWSH01005248 PCG64337.1 ODYU01002903 SOQ40975.1 PCG64338.1 KZ150575 PZC70500.1 GANO01003282 JAB56589.1 KQ459581 KPI99106.1 ODYU01004086 SOQ43592.1 NWSH01000257 PCG77965.1 MH200930 AXS59140.1 KPJ17583.1 PZC71292.1 KZ150324 PZC71401.1 NWSH01000008 PCG80945.1 AY672794 AAV91016.1 KQ460130 KPJ17530.1 NWSH01002560 PCG67981.1 KPJ02244.1 JTDY01004329 KOB68290.1 KZ150603 KZ150581 PZC70467.1 PZC70496.1 BABH01036383 BABH01036384 BABH01036385 BABH01036386 BABH01036387 BABH01036388 BABH01036389 BABH01036390 BABH01036391 KZ150249 PZC71840.1 KZ150503 PZC70659.1 JF431073 AEB21638.1 KZ150642 PZC70421.1 SOQ43594.1 PZC70465.1 NWSH01000265 PCG77882.1 KZ150429 PZC70909.1 NWSH01000060 PCG80065.1 AY672798 AAV91020.1 ODYU01003947 SOQ43326.1 ODYU01008592 SOQ52317.1 PCG72936.1 KZ150531 PZC70597.1 ODYU01010076 SOQ54921.1 SOQ39557.1 PCG77964.1 ODYU01004095 SOQ43606.1 ODYU01001487 SOQ37758.1 APCN01003350 AAAB01008984 EAL38946.3 KZ150189 PZC72424.1 ODYU01006166 SOQ47783.1 PCG80944.1 KZ150652 PZC70412.1 JTDY01003277 KOB69816.1 NWSH01003312 PCG66586.1 AK402054 BAM18676.1 ATLV01018364 KE525231 KFB42999.1 AXCN02000734 KZ149920 PZC77778.1 DS234510 EDS34246.1 AXCM01000044 PCG80941.1 UFQS01000184 UFQT01000184 SSX00849.1 SSX21229.1 KPJ02243.1 CH477825 EAT35883.2 PCG80964.1 GGFJ01008830 MBW57971.1 DS231821 EDS44979.1 ADMH02000599 ETN65690.1 KZ150408 PZC70992.1 GFDL01005857 JAV29188.1 PZC72431.1 PZC70661.1 PZC70910.1 SOQ37759.1 AJWK01025808
BABH01041623 BABH01041624 KZ150484 PZC70714.1 NWSH01003268 PCG66648.1 NWSH01001045 PCG72935.1 KZ150278 PZC71645.1 GAIX01012077 JAA80483.1 KZ150344 PZC71293.1 PCG66646.1 RSAL01000491 RVE41548.1 KQ460127 KPJ17576.1 ODYU01000940 SOQ36457.1 ODYU01002282 SOQ39556.1 PZC71287.1 AY672797 AAV91019.1 KQ459249 KPJ02240.1 NWSH01005248 PCG64337.1 ODYU01002903 SOQ40975.1 PCG64338.1 KZ150575 PZC70500.1 GANO01003282 JAB56589.1 KQ459581 KPI99106.1 ODYU01004086 SOQ43592.1 NWSH01000257 PCG77965.1 MH200930 AXS59140.1 KPJ17583.1 PZC71292.1 KZ150324 PZC71401.1 NWSH01000008 PCG80945.1 AY672794 AAV91016.1 KQ460130 KPJ17530.1 NWSH01002560 PCG67981.1 KPJ02244.1 JTDY01004329 KOB68290.1 KZ150603 KZ150581 PZC70467.1 PZC70496.1 BABH01036383 BABH01036384 BABH01036385 BABH01036386 BABH01036387 BABH01036388 BABH01036389 BABH01036390 BABH01036391 KZ150249 PZC71840.1 KZ150503 PZC70659.1 JF431073 AEB21638.1 KZ150642 PZC70421.1 SOQ43594.1 PZC70465.1 NWSH01000265 PCG77882.1 KZ150429 PZC70909.1 NWSH01000060 PCG80065.1 AY672798 AAV91020.1 ODYU01003947 SOQ43326.1 ODYU01008592 SOQ52317.1 PCG72936.1 KZ150531 PZC70597.1 ODYU01010076 SOQ54921.1 SOQ39557.1 PCG77964.1 ODYU01004095 SOQ43606.1 ODYU01001487 SOQ37758.1 APCN01003350 AAAB01008984 EAL38946.3 KZ150189 PZC72424.1 ODYU01006166 SOQ47783.1 PCG80944.1 KZ150652 PZC70412.1 JTDY01003277 KOB69816.1 NWSH01003312 PCG66586.1 AK402054 BAM18676.1 ATLV01018364 KE525231 KFB42999.1 AXCN02000734 KZ149920 PZC77778.1 DS234510 EDS34246.1 AXCM01000044 PCG80941.1 UFQS01000184 UFQT01000184 SSX00849.1 SSX21229.1 KPJ02243.1 CH477825 EAT35883.2 PCG80964.1 GGFJ01008830 MBW57971.1 DS231821 EDS44979.1 ADMH02000599 ETN65690.1 KZ150408 PZC70992.1 GFDL01005857 JAV29188.1 PZC72431.1 PZC70661.1 PZC70910.1 SOQ37759.1 AJWK01025808
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000037510
+ More
UP000075901 UP000076407 UP000075840 UP000007062 UP000075882 UP000075902 UP000075903 UP000075900 UP000030765 UP000076408 UP000075880 UP000075886 UP000002320 UP000075883 UP000075881 UP000075884 UP000008820 UP000000673 UP000075920 UP000069272 UP000092461
UP000075901 UP000076407 UP000075840 UP000007062 UP000075882 UP000075902 UP000075903 UP000075900 UP000030765 UP000076408 UP000075880 UP000075886 UP000002320 UP000075883 UP000075881 UP000075884 UP000008820 UP000000673 UP000075920 UP000069272 UP000092461
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JLA2
H9JLA5
H9JY18
A0A2W1B9D9
A0A2A4J4P7
A0A2A4JLI0
+ More
A0A2W1BCP3 S4NXP2 A0A2W1BHZ8 A0A2A4J4I5 A0A3S2NPZ4 A0A0N1IHL2 A0A2H1V6L6 A0A2H1VFJ7 A0A2W1BD72 Q5MPB3 A0A194QB71 A0A2A4IWJ7 A0A2H1VJG7 A0A2A4IY52 A0A2W1BC76 U5ESE3 A0A194Q0M3 A0A2H1VSY3 A0A2A4K113 A0A346RAE0 A0A0N1PHF7 A0A2W1B4D5 A0A2W1B9E2 A0A2A4K9I3 Q5MPB6 A0A0N0PDL6 A0A2A4J7N6 A0A194Q9Z8 A0A0L7KYR5 A0A2W1BAK2 H9JJA9 A0A2W1BGC1 A0A2W1B0L2 F5BZV1 A0A2W1B4D4 A0A2H1VTL8 A0A2W1B6N2 A0A2A4K2L1 A0A2W1B2E4 A0A2A4K714 Q5MPB2 A0A2H1VRC0 A0A2H1WH10 A0A2A4JMC3 A0A2W1BB28 A0A2H1WPC1 A0A2H1VFF0 A0A2A4K236 A0A2H1VS51 A0A182SUQ0 A0A2H1VA93 A0A182XUQ8 A0A182IIN2 Q5TNC5 A0A182KNG1 A0A2W1BFG9 A0A182THH3 A0A2H1W424 A0A182VNZ6 A0A2A4KAK6 A0A2W1AYZ6 A0A0L7L3B8 A0A2A4J4C8 I4DL86 A0A182S507 A0A084VYF5 A0A182Y982 A0A182JES5 A0A182Q673 A0A2W1BRV9 B0XLJ6 A0A182MQJ3 A0A2A4K9N2 A0A182JZN4 A0A336LWU0 A0A194Q9Q9 A0A182NZ25 Q16NE9 A0A2A4KAM6 A0A2M4BYH7 B0W1A9 W5JRY9 A0A182WR24 A0A182FZY7 A0A2W1B2N2 A0A1Q3FNX1 A0A2W1BDH3 A0A2W1B5J0 A0A2W1BDD3 A0A2H1VA97 A0A1B0CS98
A0A2W1BCP3 S4NXP2 A0A2W1BHZ8 A0A2A4J4I5 A0A3S2NPZ4 A0A0N1IHL2 A0A2H1V6L6 A0A2H1VFJ7 A0A2W1BD72 Q5MPB3 A0A194QB71 A0A2A4IWJ7 A0A2H1VJG7 A0A2A4IY52 A0A2W1BC76 U5ESE3 A0A194Q0M3 A0A2H1VSY3 A0A2A4K113 A0A346RAE0 A0A0N1PHF7 A0A2W1B4D5 A0A2W1B9E2 A0A2A4K9I3 Q5MPB6 A0A0N0PDL6 A0A2A4J7N6 A0A194Q9Z8 A0A0L7KYR5 A0A2W1BAK2 H9JJA9 A0A2W1BGC1 A0A2W1B0L2 F5BZV1 A0A2W1B4D4 A0A2H1VTL8 A0A2W1B6N2 A0A2A4K2L1 A0A2W1B2E4 A0A2A4K714 Q5MPB2 A0A2H1VRC0 A0A2H1WH10 A0A2A4JMC3 A0A2W1BB28 A0A2H1WPC1 A0A2H1VFF0 A0A2A4K236 A0A2H1VS51 A0A182SUQ0 A0A2H1VA93 A0A182XUQ8 A0A182IIN2 Q5TNC5 A0A182KNG1 A0A2W1BFG9 A0A182THH3 A0A2H1W424 A0A182VNZ6 A0A2A4KAK6 A0A2W1AYZ6 A0A0L7L3B8 A0A2A4J4C8 I4DL86 A0A182S507 A0A084VYF5 A0A182Y982 A0A182JES5 A0A182Q673 A0A2W1BRV9 B0XLJ6 A0A182MQJ3 A0A2A4K9N2 A0A182JZN4 A0A336LWU0 A0A194Q9Q9 A0A182NZ25 Q16NE9 A0A2A4KAM6 A0A2M4BYH7 B0W1A9 W5JRY9 A0A182WR24 A0A182FZY7 A0A2W1B2N2 A0A1Q3FNX1 A0A2W1BDH3 A0A2W1B5J0 A0A2W1BDD3 A0A2H1VA97 A0A1B0CS98
PDB
4D7G
E-value=0.000283298,
Score=98
Ontologies
GO
Topology
Length:
88
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.18898
outside
1 - 88
Population Genetic Test Statistics
Pi
204.934131
Theta
167.334747
Tajima's D
-1.340812
CLR
0.665213
CSRT
0.0826958652067397
Interpretation
Uncertain
