Gene
KWMTBOMO06943
Pre Gene Modal
BGIBMGA014438
Annotation
PREDICTED:_uncharacterized_protein_LOC105841867_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.073 PlasmaMembrane Reliability : 1.553
Sequence
CDS
ATGGCATGGGGTGTTAAAGAAAATAGGGGGGACTACTGCGTGGTCGGCACCAACGAACCCACCCACGTTCCAACGTTGGCCCGGTACCAACCGGACGTTTTGGACGTGTGTGTTCACAAGGGGCTACGGTGCCCTCTAGCGATAGAGGCACTGTACGACCTGAGTACCCCTCACCTACCGATCTTGGTCACGGTGGGTTTGGGGCTCACTCGGGTCGCGACATCCCCTCTCAGGCCTAGGGTCGACTGGCAGTCCTTCACGGGACTGCTGGCCGACCAATCCCTGTTTCAAGACCCGTCTACCCCTGATGAGGTCGACACGGCGGCCTCCCGTCTCGTCGACACCATCAGGGGAGCCCTGGACCAAGCGACGACTCTGGTACCCAGCGGGGGGCCCAGAGCGGAACTCCCGGTGCACCTCAAGACATTAATTCGTCGGAAGAGGGCACTGAGGAGGCTCTGGGCGACATCTAGGTGTCCCAGGATTAAGCGCGAGCTAAACCGACTTGAGGATGAGCTGGCGACTAAGATGCGGACTTACCGGGGTGACTCCTGGGAGGGGGGACTGTTGGACGCGTCCGATGACCGTACGATGGCCCCCCTTCACCGCATCTGTCGCCGGCTCACAAACAAGCCTTCCCCCACTTGCCCCTTGGAGGACGAGGGGGGAAGACGGTGTTTTACACCCTTGGCTCGTGCTGAAGTCCTGGCCAACTCGCTGGAGCGGCAGTTCACTCCGAATGTCTCTGCACCCTCGATGGTTGCATTCCATCGGGAGGTGTCAGAGGGCGTAATCCAATTCCTCAAACCGGAATCGCCGGGAGTAGAGCTGGGAGATGACGACTTAGTCACTCCTAGCGAGGTGCGCCTCCTCATCAAGCTGATGAAGAGGCGCAAAGCCCCCGGCGAGGACGGTATTCCTCCCCTCGCCTTGCAAAACCTCCCTCCCTCAATCGTGACACTTCCCTGGAAGATCATCGTCTTGCCCAAACCCGGCAAGGACAGGAGAAAGCCCTCCAGTTACCGACCGATTACCTTGCTCTCGCACGTGGGCAAGTTGTTCGAGCGGCTGCTCCTGAGGAGAATATCGCCGTATATTCTCCTCAGGCTGGAGCAATTCGGCTTTAGAAGCGGCCACTCGACGACTCTGCAACTGACCCGTATCCTGCATCACTTGGCGGACCGGCGAAACTCGGGCCAATACACGGTCGCGGTCCTTCTCGATATCGAGAAGGCCTTCGACCGTGTGTGGCACGAGGGGCTGGTCCACAAGCTGCGGGACGCGGGCCTACCGCACGCTCACGTGCGACTGCTCGGGTCGTACCTGGAGGGCAGAACCTTCTTTGTCGCGGTCGAGGGCGCACGGTCTTCCGTGCGTCCCATTACGGCCGGGGTTCCCCAGGGTAGTGTCCTATCACCTATCCTATACGCCCTCTACACCAATGACATCCCCACCCTGGAGGGTCACCTTCGGGCGTGGAAGGAGGACGTGAGGTTGGCCTTGTTCGCCGACGACTCGGCCTACTTCTCGTCCTCTCCGTTCCCCTCTGCAGCCATTATGAGGATGCTTTTGGATTTACTGCCCCAGTGGCTGGACCGTTGGAGAGTCGCGGTCAATGTGGGAAAGACCGCGGCCTTACTCTACGGTCCGGTCCGTATCAGAGTCGTCCCAAAGAAACTCAGCCTCCGAGGTGTGAATGTCGAGTTCAGGACCACGGTCCGGTATCTCGGATGCGACATCGACCGCTCCTTGAGCATGGTCAGGGCGGTTCGTGGATACCGCACCGTCTCGTTTGAGGCGGCGTGCGTGCTAGCTGGGACGCCTCCCTGGGACCTAGAAGCGGAGGCGCTCGCTGCGGATTACGCGTGGCGATGCGATCTCCGCTCCAGGGGGGAGCCGCGTCCCGGCGCGGCGGAAATTCGAGCGCGGAAGCTTCAATCTCGGCGTGCCGTGCTCGAGGCGTGGTCTCGCCGCCTGGCGGACCCCGCCTACGGGCGACGGACTGTCGAGGCGATCCGCCCGGTCCTCTCAGACTGGGTGAATCGCGACCGAGGACGTCTCACCTTCCGAGCGACGCAGGTGCTCACGGGACACGGCTGTTTCGGTCGCTACCTGCACCTCGTCGCCCGGAGGGAGCCGACGCCGAAGTGCCACCACTGCAGTGGCTGCAACGAGGACACGGCGGAGCACACGCTCGCGTACTGCACCGCTTTCGCGGAGCAGCGCCGCGTCCTCGTTGCAAAAATAGGACCGGACTTGTCGCTTCCGACCGTCGTGGCTACGATGCTCGGCAGCGACGAGTCCTGGCAGGCGATGCTCGACTTCTGCGAGTCCACCAACTCGCAGAAGGAGGCGGCGGAACGGGAGAGGGAGAGCTCTTCTTCCCTCTCGGCGCCGTGCCGCCGCCGTCGAGCCGGGGTTCGGAGGAGGGCGTTTGTCCAGCTCCAGCCCCTTTGTGTTTACGAGGCAGTCACCATGAGGCGATCGACAAAAGTCATCTCGCTGGTCAACCAGTCAGATAAGGGAGAAATCAAAACATGTTCCAGATTCATGAAGATATACTTCTTCGTGATTTACATTCTTACTGGCTTCAACTTCGGTTTCTACACCGGCTGCGGCCTCAACTTCTTGCGAGTCATACAAGCTTCAGTCTTATTACTGAGATTAAGCGTCGCTTCATACAGCATGTATATTGCACGTTATTCCCCGCTTCTGGAAGTTATCTGGTGCTGTTTGACTGCCAGTGAAAACTTAGCGGTAGTGGTGTGCTTCATGCTGAGTAGATCTGCTCTGAGCTGCAAGAACCTGTTTGAGTACTTGTACAGTGTCGACCAGGAGTTGAAGAAGAGTGTCGGTCCCAGCATCGAAGTGAAGCTCGCGCTGTACACGGTCGTGGTGAGCGTGCTGAGACTGATTATCTACGTGTTCTGTGCTACCGCCTACTACAGAAAACTGTTCGATGGACTTCGCCTCGAGTTGCTCTACCACACACCTTGTTACTCATTGGATCTATATCTCGTGGTACACTTTACAATATTTCATTCGGTTTACTGTCGACTGAAGGCGCTGAGAATATCTCTGAACGAGAAATTTGATGTTTACAAAGGTACTCTCATATACAAGTCGTTGATTGATAATTTGGAAGAAATCAAGAAAAGTTTGGATGTTCCGGTAAATATTTATAGCTCGGTTTCAGTAGTTTTAATTTGCTGA
Protein
MAWGVKENRGDYCVVGTNEPTHVPTLARYQPDVLDVCVHKGLRCPLAIEALYDLSTPHLPILVTVGLGLTRVATSPLRPRVDWQSFTGLLADQSLFQDPSTPDEVDTAASRLVDTIRGALDQATTLVPSGGPRAELPVHLKTLIRRKRALRRLWATSRCPRIKRELNRLEDELATKMRTYRGDSWEGGLLDASDDRTMAPLHRICRRLTNKPSPTCPLEDEGGRRCFTPLARAEVLANSLERQFTPNVSAPSMVAFHREVSEGVIQFLKPESPGVELGDDDLVTPSEVRLLIKLMKRRKAPGEDGIPPLALQNLPPSIVTLPWKIIVLPKPGKDRRKPSSYRPITLLSHVGKLFERLLLRRISPYILLRLEQFGFRSGHSTTLQLTRILHHLADRRNSGQYTVAVLLDIEKAFDRVWHEGLVHKLRDAGLPHAHVRLLGSYLEGRTFFVAVEGARSSVRPITAGVPQGSVLSPILYALYTNDIPTLEGHLRAWKEDVRLALFADDSAYFSSSPFPSAAIMRMLLDLLPQWLDRWRVAVNVGKTAALLYGPVRIRVVPKKLSLRGVNVEFRTTVRYLGCDIDRSLSMVRAVRGYRTVSFEAACVLAGTPPWDLEAEALAADYAWRCDLRSRGEPRPGAAEIRARKLQSRRAVLEAWSRRLADPAYGRRTVEAIRPVLSDWVNRDRGRLTFRATQVLTGHGCFGRYLHLVARREPTPKCHHCSGCNEDTAEHTLAYCTAFAEQRRVLVAKIGPDLSLPTVVATMLGSDESWQAMLDFCESTNSQKEAAERERESSSSLSAPCRRRRAGVRRRAFVQLQPLCVYEAVTMRRSTKVISLVNQSDKGEIKTCSRFMKIYFFVIYILTGFNFGFYTGCGLNFLRVIQASVLLLRLSVASYSMYIARYSPLLEVIWCCLTASENLAVVVCFMLSRSALSCKNLFEYLYSVDQELKKSVGPSIEVKLALYTVVVSVLRLIIYVFCATAYYRKLFDGLRLELLYHTPCYSLDLYLVVHFTIFHSVYCRLKALRISLNEKFDVYKGTLIYKSLIDNLEEIKKSLDVPVNIYSSVSVVLIC
Summary
Uniprot
ProteinModelPortal
PDB
6AR3
E-value=7.25672e-06,
Score=123
Ontologies
GO
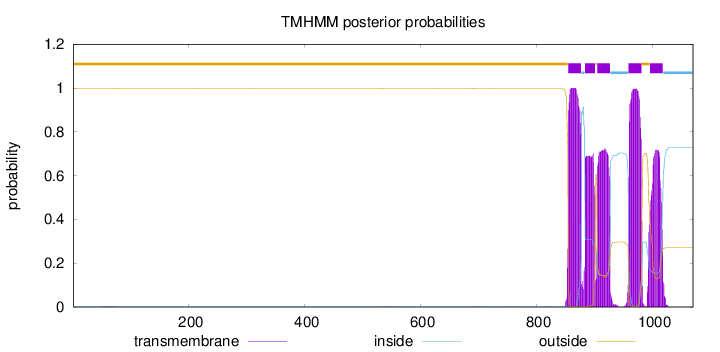
Topology
Length:
1070
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
89.6336799999999
Exp number, first 60 AAs:
0.02555
Total prob of N-in:
0.00339
outside
1 - 854
TMhelix
855 - 877
inside
878 - 883
TMhelix
884 - 901
outside
902 - 904
TMhelix
905 - 927
inside
928 - 958
TMhelix
959 - 981
outside
982 - 995
TMhelix
996 - 1018
inside
1019 - 1070
Population Genetic Test Statistics
Pi
214.109893
Theta
166.452343
Tajima's D
1.417245
CLR
0
CSRT
0.769461526923654
Interpretation
Uncertain
