Gene
KWMTBOMO06941
Pre Gene Modal
BGIBMGA014434
Annotation
PREDICTED:_proton-coupled_folate_transporter-like_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.925
Sequence
CDS
ATGCTGGATGAAACAGTAGTTCCAGAGAGCGAGTCTGACGTGATGGTGCGGAGAGCGAGGAGACCTGGCAGTACAATCGAGCTGTCATTCTTCATGACAACGCTGGGTTTGTCGCTTACCGGTGTCGCGATAAGCAACTTAATATTATACAGAACGTGTGTCCACGTGTTGCGGCACGGCCGGGCCGAGTGCTCGTCGTTCCTGGCGGTGGACAAGACTAACGACACGCAGTACTTAGAGAAGGAGGTGCAGAGGTACGCGATCTTCGTGAACAGTACAAAGAGCGTGCTCGAGTCCCTGGCACCGGCCGTACTGAGTCTCTTCCTGGGCTCGTGGTCCGACCGACACGGAAGGAAGCCGCTCATTGTGTGGCCTCTTTTTGGGATCGCAACTACTTCAATGTTGGTACTGATATATAGTATGGTTGATGTGGGACCCTGGTGGTTCATTCTGACGTCCGTACCATACTCGCTGGCTGGCGGAGTGGCCACCCTGTCCACGGGGTCTTACTCCTACATGAGTGACGTCACGAGCACTGAGAAACGAACCGCAAGAATGTTCACGGTGCAGCTCGCAGCGTCAGGCGGCGTGGTGGCGGGCTCGCTGCTCAGTGCGCCGGTGCTGCGCGCGGTGGGCGCCGTGTACCTGCTGCTGGCCGGGGCCTCCGTCGCGGCCTTTACTTATGCCTTCACGAATGTGTTCTTGAATGAATCGTTAACAGGAGTCGTTCAGGGCAGCATGACCTCGATAATGGACCTGCAGCTGGTGACAGACATGAGCAGGGAGTGCTTCAGACGCCGCCCGAACAACGGCAGGGCTCAGGTGCTCGTCGTCGCAGCCGCCTCGAGCCTCTCCGCCTTCGTTACGTTTGGGATCATGAATTTGGAATACTTGTATACCAGAGAGAAACTGCACTGGACCCTCAGCGAGTTTACCGTGTACTCGGCTGTAAACACTCTGTTGAGTTTCTTTGGAGGGCTCATAGGAATGTCGGTACTTCAGGGACTGTTCAAGATCAGCGACTTGCCGCTGGCAGCTGTTGCATTCGTCTCTGCTGCAGCGGAATATGTTGCGAAAACTTTAGCAGTTGATTCATGGCATATGTATTTGGGTGCTAGTTTAGCCATTTTCAAAGGTTTGTCAGCGTCGTTGCTCCGATCTTTCGTGAGTAAGGTCCTCCCTGCCGATGTGGTGACAAAGTTCCTTGCGTTGATGTCCGCCGTGGAGAGCCTGTGCCCGCTGCTGGCGCCCCTGGTCTACAGCACCCTGTACGGAGCCACCGTGTCCACCGCGCCAGCCTCCGTCTACGTGCTCAGCACGGGAATTATTGTTTTTTGCATTGTATTAATAGGAGTAGTTTTGTATTTCCGAAGACATGCTACGACACCTTATGAAGTACTACAACCAGAGCCCTTCTGA
Protein
MLDETVVPESESDVMVRRARRPGSTIELSFFMTTLGLSLTGVAISNLILYRTCVHVLRHGRAECSSFLAVDKTNDTQYLEKEVQRYAIFVNSTKSVLESLAPAVLSLFLGSWSDRHGRKPLIVWPLFGIATTSMLVLIYSMVDVGPWWFILTSVPYSLAGGVATLSTGSYSYMSDVTSTEKRTARMFTVQLAASGGVVAGSLLSAPVLRAVGAVYLLLAGASVAAFTYAFTNVFLNESLTGVVQGSMTSIMDLQLVTDMSRECFRRRPNNGRAQVLVVAAASSLSAFVTFGIMNLEYLYTREKLHWTLSEFTVYSAVNTLLSFFGGLIGMSVLQGLFKISDLPLAAVAFVSAAAEYVAKTLAVDSWHMYLGASLAIFKGLSASLLRSFVSKVLPADVVTKFLALMSAVESLCPLLAPLVYSTLYGATVSTAPASVYVLSTGIIVFCIVLIGVVLYFRRHATTPYEVLQPEPF
Summary
Uniprot
H9JY16
A0A2H1W9A6
A0A3S2NSY9
A0A212FJR3
A0A2W1BW92
A0A2A4JNQ2
+ More
A0A194QJP3 A0A194R1C0 A0A194QXF5 A0A194QK93 A0A212FJP0 I4DLJ4 A0A2J7QN19 A0A2J7QN30 A0A1A9WMU3 A0A1I8MY58 T1PBL8 A0A1W4UK69 A0A3B0JLW6 A0A1I8Q8R6 B4NNL7 Q961H2 A0A0J9R8V6 A0A1A9VPF0 B4HSN3 A0A1D6ZLZ8 B4QGT0 A0A034VGW4 A0A0K8VG66 A0A1B0F9V5 B4P3Y3 W8BDV4 Q28XW4 A0A0A1WK53 B4NNL5 B3N7K3 A0A0L0CFV4 B4KQL2 A0A0K8W9V2 B3MDH2 A0A336KPD5 A0A336MDC3 A0A182G7A7 A0A0K8WFD3 B4ME54 A0A182G671 A0A2M3Z2G2 A0A0K8UXE3 A0A0K8VZ06 A0A034VFP1 A0A2M3Z2I0 A0A2M4BLZ1 B0X701 A0A0K8VTP2 A0A0T6B6S1 Q1EC10 B4QGT1 A0A0A1XC01 A0A084W2H8 B3MDH1 W8BM07 A0A2M4APQ5 A0A0J9R8S1 A0A1W4UXN0 W8B4I3 A0A1S4J131 A0A182TA87 A0A1S4EZI1 Q17KH2 A0A2M4BND9 A0A0A1XF06 A0A2M4APY0 A0A1Q3FGC4 A0A1Q3FGF0 B3N7K2 A0A336MM13 B4P3Y4 W5JNI9 A0A0M4E5K0 A0A1L8DE75 A0A1L8DEH4 A0A1L8DE79 A0A1L8DE68 Q28XW3 A0A1Q3FSP6 A0A182PGG7 A0A182RA17 B4GH50 A0A139WEK6 A0A182YFN8 A0A3F2YSU9 A0A182JQA3 Q7Q7G7 A0A182N879 A0A182UEH6 A0A182MM54 A0A1B0CSD9 A0A0K8W010
A0A194QJP3 A0A194R1C0 A0A194QXF5 A0A194QK93 A0A212FJP0 I4DLJ4 A0A2J7QN19 A0A2J7QN30 A0A1A9WMU3 A0A1I8MY58 T1PBL8 A0A1W4UK69 A0A3B0JLW6 A0A1I8Q8R6 B4NNL7 Q961H2 A0A0J9R8V6 A0A1A9VPF0 B4HSN3 A0A1D6ZLZ8 B4QGT0 A0A034VGW4 A0A0K8VG66 A0A1B0F9V5 B4P3Y3 W8BDV4 Q28XW4 A0A0A1WK53 B4NNL5 B3N7K3 A0A0L0CFV4 B4KQL2 A0A0K8W9V2 B3MDH2 A0A336KPD5 A0A336MDC3 A0A182G7A7 A0A0K8WFD3 B4ME54 A0A182G671 A0A2M3Z2G2 A0A0K8UXE3 A0A0K8VZ06 A0A034VFP1 A0A2M3Z2I0 A0A2M4BLZ1 B0X701 A0A0K8VTP2 A0A0T6B6S1 Q1EC10 B4QGT1 A0A0A1XC01 A0A084W2H8 B3MDH1 W8BM07 A0A2M4APQ5 A0A0J9R8S1 A0A1W4UXN0 W8B4I3 A0A1S4J131 A0A182TA87 A0A1S4EZI1 Q17KH2 A0A2M4BND9 A0A0A1XF06 A0A2M4APY0 A0A1Q3FGC4 A0A1Q3FGF0 B3N7K2 A0A336MM13 B4P3Y4 W5JNI9 A0A0M4E5K0 A0A1L8DE75 A0A1L8DEH4 A0A1L8DE79 A0A1L8DE68 Q28XW3 A0A1Q3FSP6 A0A182PGG7 A0A182RA17 B4GH50 A0A139WEK6 A0A182YFN8 A0A3F2YSU9 A0A182JQA3 Q7Q7G7 A0A182N879 A0A182UEH6 A0A182MM54 A0A1B0CSD9 A0A0K8W010
Pubmed
19121390
22118469
28756777
26354079
22651552
25315136
+ More
17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 17550304 24495485 15632085 25830018 26108605 26483478 24438588 17510324 20920257 23761445 18362917 19820115 25244985 12364791 14747013 17210077
17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 17550304 24495485 15632085 25830018 26108605 26483478 24438588 17510324 20920257 23761445 18362917 19820115 25244985 12364791 14747013 17210077
EMBL
BABH01041627
ODYU01007151
SOQ49679.1
RSAL01000235
RVE43765.1
AGBW02008244
+ More
OWR53966.1 KZ149917 PZC77915.1 NWSH01001023 PCG73050.1 KQ458575 KPJ05793.1 KQ461073 KPJ09641.1 KPJ09640.1 KPJ05794.1 OWR53967.1 AK402162 BAM18784.1 NEVH01013201 PNF29985.1 PNF29986.1 KA645505 AFP60134.1 OUUW01000001 SPP74236.1 CH964282 EDW85956.1 AY051590 AE013599 AAK93014.1 AAM68813.1 CM002911 KMY92466.1 CH480816 EDW47060.1 KT722703 ANS71638.1 CM000362 EDX06305.1 GAKP01017238 JAC41714.1 GDHF01014395 GDHF01002448 JAI37919.1 JAI49866.1 CCAG010014464 CM000157 EDW89466.1 GAMC01009658 JAB96897.1 CM000071 EAL26202.1 GBXI01015402 JAC98889.1 EDW85954.1 CH954177 EDV59408.1 JRES01000440 KNC31125.1 CH933808 EDW08181.1 GDHF01004457 JAI47857.1 CH902619 EDV36420.1 UFQS01000553 UFQT01000553 SSX04853.1 SSX25216.1 UFQT01000972 SSX28236.1 JXUM01008009 JXUM01008010 KQ560251 KXJ83555.1 GDHF01002438 JAI49876.1 CH940662 EDW58819.1 JXUM01007622 JXUM01007623 KQ560244 KXJ83610.1 GGFM01001956 MBW22707.1 GDHF01020977 JAI31337.1 GDHF01008166 JAI44148.1 GAKP01016836 JAC42116.1 GGFM01001978 MBW22729.1 GGFJ01004822 MBW53963.1 DS232431 EDS41672.1 GDHF01010070 JAI42244.1 LJIG01009453 KRT83049.1 BT025924 AAM68814.2 ABG02168.1 EDX06306.1 GBXI01005820 JAD08472.1 ATLV01019619 KE525275 KFB44422.1 EDV36419.2 GAMC01008532 JAB98023.1 GGFK01009429 MBW42750.1 KMY92467.1 GAMC01013083 JAB93472.1 CH477224 EAT47179.1 GGFJ01005449 MBW54590.1 GBXI01004318 JAD09974.1 GGFK01009524 MBW42845.1 GFDL01008438 JAV26607.1 GFDL01008415 JAV26630.1 EDV59407.2 UFQS01001699 UFQT01001699 SSX11900.1 SSX31462.1 EDW89467.2 ADMH02000550 ETN65922.1 CP012524 ALC41566.1 GFDF01009323 JAV04761.1 GFDF01009324 JAV04760.1 GFDF01009322 JAV04762.1 GFDF01009321 JAV04763.1 EAL26203.1 GFDL01004431 JAV30614.1 CH479183 EDW35820.1 KQ971354 KYB26390.1 APCN01004443 AAAB01008960 EAA11797.4 AXCM01004465 AJWK01025916 GDHF01008134 JAI44180.1
OWR53966.1 KZ149917 PZC77915.1 NWSH01001023 PCG73050.1 KQ458575 KPJ05793.1 KQ461073 KPJ09641.1 KPJ09640.1 KPJ05794.1 OWR53967.1 AK402162 BAM18784.1 NEVH01013201 PNF29985.1 PNF29986.1 KA645505 AFP60134.1 OUUW01000001 SPP74236.1 CH964282 EDW85956.1 AY051590 AE013599 AAK93014.1 AAM68813.1 CM002911 KMY92466.1 CH480816 EDW47060.1 KT722703 ANS71638.1 CM000362 EDX06305.1 GAKP01017238 JAC41714.1 GDHF01014395 GDHF01002448 JAI37919.1 JAI49866.1 CCAG010014464 CM000157 EDW89466.1 GAMC01009658 JAB96897.1 CM000071 EAL26202.1 GBXI01015402 JAC98889.1 EDW85954.1 CH954177 EDV59408.1 JRES01000440 KNC31125.1 CH933808 EDW08181.1 GDHF01004457 JAI47857.1 CH902619 EDV36420.1 UFQS01000553 UFQT01000553 SSX04853.1 SSX25216.1 UFQT01000972 SSX28236.1 JXUM01008009 JXUM01008010 KQ560251 KXJ83555.1 GDHF01002438 JAI49876.1 CH940662 EDW58819.1 JXUM01007622 JXUM01007623 KQ560244 KXJ83610.1 GGFM01001956 MBW22707.1 GDHF01020977 JAI31337.1 GDHF01008166 JAI44148.1 GAKP01016836 JAC42116.1 GGFM01001978 MBW22729.1 GGFJ01004822 MBW53963.1 DS232431 EDS41672.1 GDHF01010070 JAI42244.1 LJIG01009453 KRT83049.1 BT025924 AAM68814.2 ABG02168.1 EDX06306.1 GBXI01005820 JAD08472.1 ATLV01019619 KE525275 KFB44422.1 EDV36419.2 GAMC01008532 JAB98023.1 GGFK01009429 MBW42750.1 KMY92467.1 GAMC01013083 JAB93472.1 CH477224 EAT47179.1 GGFJ01005449 MBW54590.1 GBXI01004318 JAD09974.1 GGFK01009524 MBW42845.1 GFDL01008438 JAV26607.1 GFDL01008415 JAV26630.1 EDV59407.2 UFQS01001699 UFQT01001699 SSX11900.1 SSX31462.1 EDW89467.2 ADMH02000550 ETN65922.1 CP012524 ALC41566.1 GFDF01009323 JAV04761.1 GFDF01009324 JAV04760.1 GFDF01009322 JAV04762.1 GFDF01009321 JAV04763.1 EAL26203.1 GFDL01004431 JAV30614.1 CH479183 EDW35820.1 KQ971354 KYB26390.1 APCN01004443 AAAB01008960 EAA11797.4 AXCM01004465 AJWK01025916 GDHF01008134 JAI44180.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000218220
UP000053268
UP000053240
+ More
UP000235965 UP000091820 UP000095301 UP000192221 UP000268350 UP000095300 UP000007798 UP000000803 UP000078200 UP000001292 UP000000304 UP000092444 UP000002282 UP000001819 UP000008711 UP000037069 UP000009192 UP000007801 UP000069940 UP000249989 UP000008792 UP000002320 UP000030765 UP000075901 UP000008820 UP000000673 UP000092553 UP000075885 UP000075900 UP000008744 UP000007266 UP000076408 UP000075840 UP000075881 UP000007062 UP000075884 UP000075902 UP000075883 UP000092461
UP000235965 UP000091820 UP000095301 UP000192221 UP000268350 UP000095300 UP000007798 UP000000803 UP000078200 UP000001292 UP000000304 UP000092444 UP000002282 UP000001819 UP000008711 UP000037069 UP000009192 UP000007801 UP000069940 UP000249989 UP000008792 UP000002320 UP000030765 UP000075901 UP000008820 UP000000673 UP000092553 UP000075885 UP000075900 UP000008744 UP000007266 UP000076408 UP000075840 UP000075881 UP000007062 UP000075884 UP000075902 UP000075883 UP000092461
Pfam
PF07690 MFS_1
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JY16
A0A2H1W9A6
A0A3S2NSY9
A0A212FJR3
A0A2W1BW92
A0A2A4JNQ2
+ More
A0A194QJP3 A0A194R1C0 A0A194QXF5 A0A194QK93 A0A212FJP0 I4DLJ4 A0A2J7QN19 A0A2J7QN30 A0A1A9WMU3 A0A1I8MY58 T1PBL8 A0A1W4UK69 A0A3B0JLW6 A0A1I8Q8R6 B4NNL7 Q961H2 A0A0J9R8V6 A0A1A9VPF0 B4HSN3 A0A1D6ZLZ8 B4QGT0 A0A034VGW4 A0A0K8VG66 A0A1B0F9V5 B4P3Y3 W8BDV4 Q28XW4 A0A0A1WK53 B4NNL5 B3N7K3 A0A0L0CFV4 B4KQL2 A0A0K8W9V2 B3MDH2 A0A336KPD5 A0A336MDC3 A0A182G7A7 A0A0K8WFD3 B4ME54 A0A182G671 A0A2M3Z2G2 A0A0K8UXE3 A0A0K8VZ06 A0A034VFP1 A0A2M3Z2I0 A0A2M4BLZ1 B0X701 A0A0K8VTP2 A0A0T6B6S1 Q1EC10 B4QGT1 A0A0A1XC01 A0A084W2H8 B3MDH1 W8BM07 A0A2M4APQ5 A0A0J9R8S1 A0A1W4UXN0 W8B4I3 A0A1S4J131 A0A182TA87 A0A1S4EZI1 Q17KH2 A0A2M4BND9 A0A0A1XF06 A0A2M4APY0 A0A1Q3FGC4 A0A1Q3FGF0 B3N7K2 A0A336MM13 B4P3Y4 W5JNI9 A0A0M4E5K0 A0A1L8DE75 A0A1L8DEH4 A0A1L8DE79 A0A1L8DE68 Q28XW3 A0A1Q3FSP6 A0A182PGG7 A0A182RA17 B4GH50 A0A139WEK6 A0A182YFN8 A0A3F2YSU9 A0A182JQA3 Q7Q7G7 A0A182N879 A0A182UEH6 A0A182MM54 A0A1B0CSD9 A0A0K8W010
A0A194QJP3 A0A194R1C0 A0A194QXF5 A0A194QK93 A0A212FJP0 I4DLJ4 A0A2J7QN19 A0A2J7QN30 A0A1A9WMU3 A0A1I8MY58 T1PBL8 A0A1W4UK69 A0A3B0JLW6 A0A1I8Q8R6 B4NNL7 Q961H2 A0A0J9R8V6 A0A1A9VPF0 B4HSN3 A0A1D6ZLZ8 B4QGT0 A0A034VGW4 A0A0K8VG66 A0A1B0F9V5 B4P3Y3 W8BDV4 Q28XW4 A0A0A1WK53 B4NNL5 B3N7K3 A0A0L0CFV4 B4KQL2 A0A0K8W9V2 B3MDH2 A0A336KPD5 A0A336MDC3 A0A182G7A7 A0A0K8WFD3 B4ME54 A0A182G671 A0A2M3Z2G2 A0A0K8UXE3 A0A0K8VZ06 A0A034VFP1 A0A2M3Z2I0 A0A2M4BLZ1 B0X701 A0A0K8VTP2 A0A0T6B6S1 Q1EC10 B4QGT1 A0A0A1XC01 A0A084W2H8 B3MDH1 W8BM07 A0A2M4APQ5 A0A0J9R8S1 A0A1W4UXN0 W8B4I3 A0A1S4J131 A0A182TA87 A0A1S4EZI1 Q17KH2 A0A2M4BND9 A0A0A1XF06 A0A2M4APY0 A0A1Q3FGC4 A0A1Q3FGF0 B3N7K2 A0A336MM13 B4P3Y4 W5JNI9 A0A0M4E5K0 A0A1L8DE75 A0A1L8DEH4 A0A1L8DE79 A0A1L8DE68 Q28XW3 A0A1Q3FSP6 A0A182PGG7 A0A182RA17 B4GH50 A0A139WEK6 A0A182YFN8 A0A3F2YSU9 A0A182JQA3 Q7Q7G7 A0A182N879 A0A182UEH6 A0A182MM54 A0A1B0CSD9 A0A0K8W010
Ontologies
GO
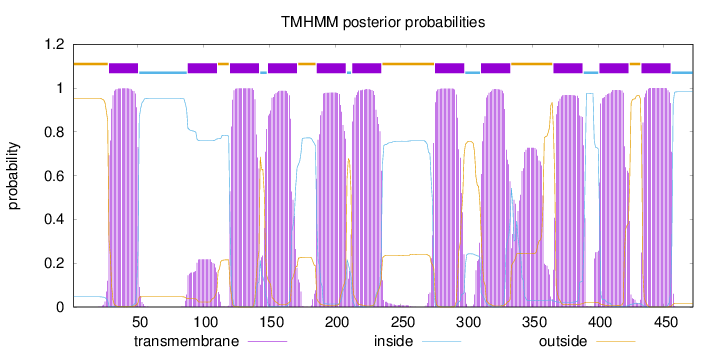
Topology
Length:
472
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
239.61297
Exp number, first 60 AAs:
22.27378
Total prob of N-in:
0.04686
POSSIBLE N-term signal
sequence
outside
1 - 27
TMhelix
28 - 50
inside
51 - 87
TMhelix
88 - 110
outside
111 - 119
TMhelix
120 - 142
inside
143 - 148
TMhelix
149 - 171
outside
172 - 185
TMhelix
186 - 208
inside
209 - 212
TMhelix
213 - 235
outside
236 - 275
TMhelix
276 - 298
inside
299 - 310
TMhelix
311 - 333
outside
334 - 365
TMhelix
366 - 388
inside
389 - 400
TMhelix
401 - 423
outside
424 - 432
TMhelix
433 - 455
inside
456 - 472
Population Genetic Test Statistics
Pi
226.63309
Theta
186.524696
Tajima's D
-2.291519
CLR
0.49762
CSRT
0.00394980250987451
Interpretation
Uncertain
