Gene
KWMTBOMO06936
Pre Gene Modal
BGIBMGA001742
Annotation
PREDICTED:_syntaxin-1B-like_[Papilio_polytes]
Location in the cell
Extracellular Reliability : 1.697
Sequence
CDS
ATGATGACCAACCTCGCGATATCCGACGAGGAGTGTCAGAGCTTGCTGGACTCGAACAACATCTCGCTGTTTGTCGACAATGTGCGCGCGGAGACGGCGGCGGCGCGGCTGGAGCTGCGCGCAGTGGAGGCGCGGCGCGAGGAGCTGGCGCGGGTCGAGGCGGCGCTGCGGGAGGTGCGCGAGCTGTTCCAGCAGCTGGCCGCGCTCGTGGCGGCGCAGCAGGACCAGATCGATAGCGTCGAGTACTACGCGCTGCAGGCCACCGAGCACGTCGAGTGCGGCGGCCAGCAACTCCTCAAGGGCACCGTCACCAGGACCAAGGCCAGAAAGAAAAAGATGAGTCTGTTGCTGTGTCTGGCGATCGGCTTCCTTATAGTACTGCTGGTCCTCTTGCTGACGTAG
Protein
MMTNLAISDEECQSLLDSNNISLFVDNVRAETAAARLELRAVEARREELARVEAALREVRELFQQLAALVAAQQDQIDSVEYYALQATEHVECGGQQLLKGTVTRTKARKKKMSLLLCLAIGFLIVLLVLLLT
Summary
Similarity
Belongs to the syntaxin family.
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF47661
SSF47661
ProteinModelPortal
Ontologies
GO
PANTHER
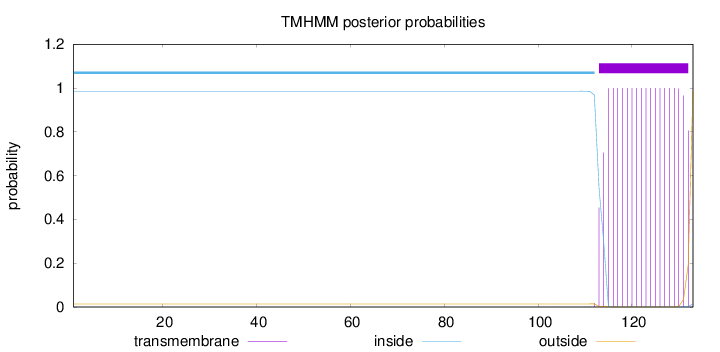
Topology
Length:
133
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.94472
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98585
inside
1 - 112
TMhelix
113 - 132
outside
133 - 133
Population Genetic Test Statistics
Pi
164.568203
Theta
65.533459
Tajima's D
0.278836
CLR
0
CSRT
0.450677466126694
Interpretation
Uncertain
