Gene
KWMTBOMO06935
Pre Gene Modal
BGIBMGA013961
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC106709759_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.063
Sequence
CDS
ATGGAATACAAAGTCCGAATTGCTAGGGCTGCAGTCAAATATTCCAATCTGGGGAGGGGTCAAGCTGCCACTCAAGAATGCCTGGCAACGGCATCCCTGGATGAATTTTCCATCGCGCTACTGCAAGAACCTTATGTTGGAAATAAAGGGATACTATCAACTAGAAACAGGATCATACAGGATAAAGGGGACAAGAATAATCCTGTCAAATCTGCCATTGTGATATTCGACAGAAGTCTGGTAGTAATTGAAAATAATTTACTCATAGATCGTAACATAGTAGCTGTTACGATAGATACACAGGGACATAGAATCGGGTGCATCTCAGTGTACTGGGAGGATTACGTGAACGTGGACACATATCTAAACAAACTGCAATACGTAATCAACAATCTAAAAACGGACACTTATATAGTTGGTGGTGATGTAAACGCTCGTAGCCAGTGGTGGGGATGCCCCAACGACGATTTAAGAGGAGCGGCTATCGCCGTTTTCTTAGCCCAGAATGATCTCCAAATACTAAACAAAGGAAACACCCCTACATTTTGGACTGTCAGACAAAACAGGCTGTACACAAGTATAATAGATATAACGGCATGCAGTGACGGACTTCTCAACTCAATATCAGATTGGAAAGTAGATGAGAATCTAATCACCACCTCCGATCATCGCTCAATCACGTTCAGCGTCGACCTTACCGGGAACAGAGGTTCTCAGAAAGGAAATCTTACCACTACAAAGATATACAATACTAATAAAGCAAACTGGCCTAAATTCAAAAATAAGCTACTGGAGCAATTCTCTGAACGAAATATCACAGTTGAAACCATCGCCTCAAAAACTGACAACGAAGAGTTCAACAAAATGGTAGAGGACTACGCGGGATGCATTGCGGTAGCCTGTGATTCCGCGATTCCTCTGATTGACCATAGTAAAAATCAATGTAGGCTATCCTGGTGGACCAAAGAACTAACCGAACTTAAAAATGAGGTTAAAAGAATCAGAAGCAAGATTAGGAAGGCAAACCCTAGAAGAAGGCCGCTTCTGATAAAAGATTATCAAGAAAGGTTAAAAGAATATAAACACACAATAATCATAACGAGGACCACCAGTTGGAAAGAATATACATCACGACAGAAGAGAGAATCGATGTGGGATAAATTCTACCGCATTATACGTACTAATAATAGAAGCAGAGATGAGGTAATGCTCAAGAAAGGGAATGACGTCCTCGACCCCCTCAAATCCGCAGAACTACTAGCTGATACATTCTATCCTAAGGACAATAAAGATATCGAAAACGAAGAACATCACCGCATACGAGAGGAGATAGCTGCATGGAGGAAGAAAATAATCGAAGAGAAGGACCCAGAATTTAAAGAAAAGCCGTTCACACCGAAAGAAATACAAATGATCTTGGACTTTGCCATGGACACTCAAAACTTCCCCAGTATTATCGAGGACGACAGTGCTAGAGAAGCGTTCCTGAAATTCGCTATAAACAGTATAAAAATTGCCAACAACAGAAATAATAGTATAATGAAATACGCTATAGATTAA
Protein
MEYKVRIARAAVKYSNLGRGQAATQECLATASLDEFSIALLQEPYVGNKGILSTRNRIIQDKGDKNNPVKSAIVIFDRSLVVIENNLLIDRNIVAVTIDTQGHRIGCISVYWEDYVNVDTYLNKLQYVINNLKTDTYIVGGDVNARSQWWGCPNDDLRGAAIAVFLAQNDLQILNKGNTPTFWTVRQNRLYTSIIDITACSDGLLNSISDWKVDENLITTSDHRSITFSVDLTGNRGSQKGNLTTTKIYNTNKANWPKFKNKLLEQFSERNITVETIASKTDNEEFNKMVEDYAGCIAVACDSAIPLIDHSKNQCRLSWWTKELTELKNEVKRIRSKIRKANPRRRPLLIKDYQERLKEYKHTIIITRTTSWKEYTSRQKRESMWDKFYRIIRTNNRSRDEVMLKKGNDVLDPLKSAELLADTFYPKDNKDIENEEHHRIREEIAAWRKKIIEEKDPEFKEKPFTPKEIQMILDFAMDTQNFPSIIEDDSAREAFLKFAINSIKIANNRNNSIMKYAID
Summary
Uniprot
Q9BLI0
Q9BLI5
Q9BLI4
Q9BLI3
A0A194QZG8
Q93139
+ More
A0A212F890 K7JMI9 A0A1B6FCA5 A0A1Y3BRS2 K7JAI1 A0A0J7KIV1 K7JWL9 A0A2R7WW78 A0A2R7WHA3 J9M8T8 A0A142LX24 T1HJW2 A0A023EX80 A0A147BCI0 A0A0J7K3F1 A0A0J7MWN5 A0A224X653 A0A1I8IBT3 A0A1S3DM51 A0A0C9QD64 J9LES5 A0A224XAL8 A0A087TI44 A0A1B6D5E4 T1I8Q1 A0A2R7WYT6 A0A1B6E1N0 A0A1B6DRM7 A0A2R7WZM4 J9KPX6 X1XKA4 A0A0K8TL06 A0A0K8TLH8 A0A1S3D5J4 A0A0J7KTH2 J9KRH3 J9JXR0 O18558 J9L0V3 V5GNF6 A0A087T6T3 A0A087TAD8 B7QBV7 A0A0K1IK29 A0A1S4EPA1 U5EW32 A0A1Y1MFF8 A0A0J7N8F6 J9LNF5 A0A0J7KF40 A0A1I8FVG9 O46184 X1X2B5 J9KMZ9
A0A212F890 K7JMI9 A0A1B6FCA5 A0A1Y3BRS2 K7JAI1 A0A0J7KIV1 K7JWL9 A0A2R7WW78 A0A2R7WHA3 J9M8T8 A0A142LX24 T1HJW2 A0A023EX80 A0A147BCI0 A0A0J7K3F1 A0A0J7MWN5 A0A224X653 A0A1I8IBT3 A0A1S3DM51 A0A0C9QD64 J9LES5 A0A224XAL8 A0A087TI44 A0A1B6D5E4 T1I8Q1 A0A2R7WYT6 A0A1B6E1N0 A0A1B6DRM7 A0A2R7WZM4 J9KPX6 X1XKA4 A0A0K8TL06 A0A0K8TLH8 A0A1S3D5J4 A0A0J7KTH2 J9KRH3 J9JXR0 O18558 J9L0V3 V5GNF6 A0A087T6T3 A0A087TAD8 B7QBV7 A0A0K1IK29 A0A1S4EPA1 U5EW32 A0A1Y1MFF8 A0A0J7N8F6 J9LNF5 A0A0J7KF40 A0A1I8FVG9 O46184 X1X2B5 J9KMZ9
Pubmed
EMBL
AB046673
BAB21516.1
AB046668
BAB21511.1
AB046669
BAB21512.1
+ More
AB046670 BAB21513.1 KQ461108 KPJ08991.1 D38414 BAA07467.1 AGBW02009775 OWR49941.1 AAZX01008070 GECZ01021958 JAS47811.1 MUJZ01008278 OTF82483.1 AAZX01006426 LBMM01006940 KMQ90177.1 AAZX01018244 KK855758 PTY23794.1 KK854804 PTY18889.1 ABLF02007471 ABLF02007472 KU543672 AMS38350.1 ACPB03028300 GBBI01004920 JAC13792.1 GEGO01007232 JAR88172.1 LBMM01015025 KMQ84953.1 LBMM01015285 KMQ84870.1 GFTR01008469 JAW07957.1 GBYB01012318 JAG82085.1 ABLF02008889 ABLF02041820 GFTR01008452 JAW07974.1 KK115320 KFM64783.1 GEDC01016405 JAS20893.1 ACPB03026099 PTY23785.1 GEDC01005462 JAS31836.1 GEDC01008950 JAS28348.1 KK856129 PTY24962.1 ABLF02019936 ABLF02057951 ABLF02011057 ABLF02048793 GDAI01002584 JAI15019.1 GDAI01002592 JAI15011.1 LBMM01003366 KMQ93606.1 ABLF02020082 ABLF02020086 ABLF02020092 ABLF02014981 U87543 AAB65093.1 ABLF02012458 ABLF02012459 GALX01005354 JAB63112.1 KK113699 KFM60822.1 KK114278 KFM62077.1 ABJB010425648 DS903648 EEC16329.1 KP771711 AKU04648.1 GANO01003141 JAB56730.1 GEZM01035254 JAV83320.1 LBMM01008384 KMQ88925.1 ABLF02006133 LBMM01008422 KMQ88894.1 U73803 AAB92393.1 ABLF02019402 ABLF02059310 ABLF02039177
AB046670 BAB21513.1 KQ461108 KPJ08991.1 D38414 BAA07467.1 AGBW02009775 OWR49941.1 AAZX01008070 GECZ01021958 JAS47811.1 MUJZ01008278 OTF82483.1 AAZX01006426 LBMM01006940 KMQ90177.1 AAZX01018244 KK855758 PTY23794.1 KK854804 PTY18889.1 ABLF02007471 ABLF02007472 KU543672 AMS38350.1 ACPB03028300 GBBI01004920 JAC13792.1 GEGO01007232 JAR88172.1 LBMM01015025 KMQ84953.1 LBMM01015285 KMQ84870.1 GFTR01008469 JAW07957.1 GBYB01012318 JAG82085.1 ABLF02008889 ABLF02041820 GFTR01008452 JAW07974.1 KK115320 KFM64783.1 GEDC01016405 JAS20893.1 ACPB03026099 PTY23785.1 GEDC01005462 JAS31836.1 GEDC01008950 JAS28348.1 KK856129 PTY24962.1 ABLF02019936 ABLF02057951 ABLF02011057 ABLF02048793 GDAI01002584 JAI15019.1 GDAI01002592 JAI15011.1 LBMM01003366 KMQ93606.1 ABLF02020082 ABLF02020086 ABLF02020092 ABLF02014981 U87543 AAB65093.1 ABLF02012458 ABLF02012459 GALX01005354 JAB63112.1 KK113699 KFM60822.1 KK114278 KFM62077.1 ABJB010425648 DS903648 EEC16329.1 KP771711 AKU04648.1 GANO01003141 JAB56730.1 GEZM01035254 JAV83320.1 LBMM01008384 KMQ88925.1 ABLF02006133 LBMM01008422 KMQ88894.1 U73803 AAB92393.1 ABLF02019402 ABLF02059310 ABLF02039177
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR012337 RNaseH-like_sf
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR031996 NVL2_nucleolin-bd
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR038100 NLV2_N_sf
IPR031961 DUF4780
IPR005312 DUF1759
IPR008737 Peptidase_asp_put
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR012337 RNaseH-like_sf
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR031996 NVL2_nucleolin-bd
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR038100 NLV2_N_sf
IPR031961 DUF4780
IPR005312 DUF1759
IPR008737 Peptidase_asp_put
Gene 3D
ProteinModelPortal
Q9BLI0
Q9BLI5
Q9BLI4
Q9BLI3
A0A194QZG8
Q93139
+ More
A0A212F890 K7JMI9 A0A1B6FCA5 A0A1Y3BRS2 K7JAI1 A0A0J7KIV1 K7JWL9 A0A2R7WW78 A0A2R7WHA3 J9M8T8 A0A142LX24 T1HJW2 A0A023EX80 A0A147BCI0 A0A0J7K3F1 A0A0J7MWN5 A0A224X653 A0A1I8IBT3 A0A1S3DM51 A0A0C9QD64 J9LES5 A0A224XAL8 A0A087TI44 A0A1B6D5E4 T1I8Q1 A0A2R7WYT6 A0A1B6E1N0 A0A1B6DRM7 A0A2R7WZM4 J9KPX6 X1XKA4 A0A0K8TL06 A0A0K8TLH8 A0A1S3D5J4 A0A0J7KTH2 J9KRH3 J9JXR0 O18558 J9L0V3 V5GNF6 A0A087T6T3 A0A087TAD8 B7QBV7 A0A0K1IK29 A0A1S4EPA1 U5EW32 A0A1Y1MFF8 A0A0J7N8F6 J9LNF5 A0A0J7KF40 A0A1I8FVG9 O46184 X1X2B5 J9KMZ9
A0A212F890 K7JMI9 A0A1B6FCA5 A0A1Y3BRS2 K7JAI1 A0A0J7KIV1 K7JWL9 A0A2R7WW78 A0A2R7WHA3 J9M8T8 A0A142LX24 T1HJW2 A0A023EX80 A0A147BCI0 A0A0J7K3F1 A0A0J7MWN5 A0A224X653 A0A1I8IBT3 A0A1S3DM51 A0A0C9QD64 J9LES5 A0A224XAL8 A0A087TI44 A0A1B6D5E4 T1I8Q1 A0A2R7WYT6 A0A1B6E1N0 A0A1B6DRM7 A0A2R7WZM4 J9KPX6 X1XKA4 A0A0K8TL06 A0A0K8TLH8 A0A1S3D5J4 A0A0J7KTH2 J9KRH3 J9JXR0 O18558 J9L0V3 V5GNF6 A0A087T6T3 A0A087TAD8 B7QBV7 A0A0K1IK29 A0A1S4EPA1 U5EW32 A0A1Y1MFF8 A0A0J7N8F6 J9LNF5 A0A0J7KF40 A0A1I8FVG9 O46184 X1X2B5 J9KMZ9
PDB
1WDU
E-value=7.09301e-36,
Score=379
Ontologies
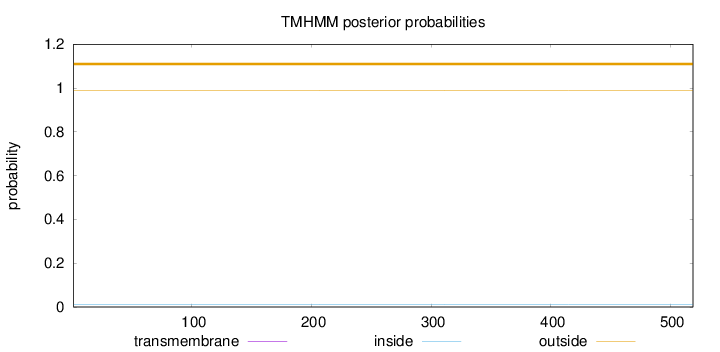
Topology
Length:
519
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00304
Exp number, first 60 AAs:
0.00043
Total prob of N-in:
0.01143
outside
1 - 519
Population Genetic Test Statistics
Pi
5.700757
Theta
152.135955
Tajima's D
-1.554823
CLR
1295.167348
CSRT
0.055597220138993
Interpretation
Uncertain
