Gene
KWMTBOMO06932 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004655
Annotation
fatty_acid_synthase_2?_partial_[Helicoverpa_assulta]
Location in the cell
Cytoplasmic Reliability : 1.086 Nuclear Reliability : 1.07
Sequence
CDS
ATGCCGAGCGTGGTGAACGGGACTAATGGCGCGGGCGTCGACGATGACATAGTATTGACCGGTCTGTCTGGGAAGCTGCCGGAGTCCAACAATATCGAAGAGTTCGCCGAGCAACTATTCGCTGGAGTGGATCTCGTCACCGCTGACAGCAGACGATGGGTCCCAGGTCTCCACGGTCTGCCAGAGAGGAATGGAAAACTTAAAGACCTCGCACACTTCGACGCCACGTTCTTCGGGGTTCACGCGAAGCAGGCAAACCTCATGGACCCGCAGCTCCGTCTTTTATTGGAGCTCACGCACGAGGCCATTATTGATGCCGGTTTCAATCCAACTGAGCTGCGCGGTTCCCGGACTGGCGTTTACGTTGGCGTCTCCAACTCTGAAACAGAAGAGATGTGGACCGCCGATCCTGACAAAATAAACGGTTACGCCCTGACTGGGTGCTGCCGTGCTATGTTCCCCAACCGCATCTCGTACACGTTCGACTTCTGCGGTCCCTCGTACGCCGTGGACACGGCCTGTTCGAGTTCAATGTTTGCGCTTGCTCAAGCCGTGACGGCGATGCGCGCGGGTCACTGCGACGCGGCCATCGTTGCCGGCACCAATCTGTGCTTGAAGCCCGCCAACTCGCTCAATTTCCACCGACTGAGCATGCTGTCTCCGCAAGGTCGTTGCGCTGCGTTTGACGCCAGCGGCAACGGTTATGTGCGGTCGGAAGCGGCCGTGGTCGTGGTCTTACAGCGCCGGGCCAACGCGCGTCGAGTGTACGCAACAGTGCGGGGCGCGAGGATGAACACAGACGGAACTAAGGTCCAGGGCATCACTTTCCCCTCGGGGGACATGCAACGCCGACTTGCGAAGGAAACTTTTGAAGAAGCCGGCTTGAGGCCGCAAGATGTAGTGTATGTTGAAGCTCACGGAACCGGTACAAAGGTCGGAGATCCACAAGAAGTGAATGCAATAGCACAGCTATTCTGCGAAAATCGTTCAGATCCTCTGCTTTTGGGCTCAGTCAAGTCAAACATGGGACATTCGGAGCCTGCTTCGGGCTTGTGCTCGATCGCTAAAGTTGTCGTCGCCATGGAGAATGGCGTCATACCTCAAAATTTACATTATAAGAGTCCAAATCTTGATATACCAGCTTTAAGCGATGGAAGAATCAAGGTAGTAGACCGGAACCTCGAATGGAACGGCGGTTTGGTGGCCGTCAACTCGTTCGGGTTCGGAGGCGCCAACGCTCACATCATCTTGGAATCGGAGCGTGTCGGCAAGCGTGTCCGCACCGAGTACGCCATACCTAGACTGGTCCTCGCTTCGGGACGCACAGAGGACGCCGTGTCCCACCTGTTGGAACACGCCGCCAAACACGCCAAGGACGCAGAGCTGCACGGTCTGCTGGACGCGATACACGCGCGGAACATCCCCGGACACGGGTACCGGGGCTACGGGCTGCTCCTGAACGAGATGCAAACTGAAATCGTCGAGACGAGCAGCGGAGATCCCAGGCCCATATGGTACGTGTTTTCTGGCATGGGGTCCCAATGGGCGGGCATGGCGAAGTGCCTGATGCGCTTGCCGGTGTTTGCGGCCAGCGTGCATCGTTCTGCCGCAGCACTAAGGCCCCATGGCATCGACCTAGTGCACGCGATCACGGAGGCCCCCGACACTGCCTTCGACGATGTCGTCATCTCATTCGTGTCGATAGCTGCCGTGCAAGTGGCCATGGTCGATGTGCTCCGCGAACTTCAGATCACTCCTGACGGCATCGTGGGACATTCCGTTGGAGAAATCGGTTGCGCGTACGCTGACGAGACGCTGACGGGCGAGCAGGCGGTGCTGGCGGCGTACTGGCGCGGGCGCAGCATCGTGGACGCGCGGCTGGACGCGGGCGCCATGGCGGCCGTGGGGCTCTCCTGGGAGCTGTGCCAGCGCCGCTGCCCGCCCGACGTGGCGCCCGCGTGCCACAACGCCAGCGACAACGTCACCATATCCGGTCCAGTTGCTTCAATAGAGAAATTCGTAGCGGAATTAACGGCCGAGGGAATCTTCGCTCGGCGGGTGAACAGCTCGGGGATCGCCTTCCACAGCAAGTACATCGCGGCCGCCGCTCCGCTGCTGAGGAAGAGCCTCGAGAAGGTCATACCTTATCCCAAGGCGCGGTCCTCTAAGTGGGTGTCCTCCTCGTTACCCAGAGACCAATGGGGATCAGAGCTGGCCAAGCTGAGCGACGTGAACTACCACGTGAACAATCTGCTGTCTCCCGTCCGCTTCGCCGACGCCCTGAAGGAGATCCCGGAGCGAGCCATCGTCGTGGAGATCGCGCCGCACGCGCTGCTGCAGGCGGTGCTGAAGCGCGCGGTGCCGGCGCCCGCGGCCGCGCACGTGGCGCTGGTGCGGCGCGACGCGGCCGACGCGCTGCTGCACCTGCTCGGCGCCGTCGGCAAGCTGTACGCGGCGGGCGCGCAGCCGCAGCCCTCCCGCCTCTACCCCGCCGTCCGCTTCCCCGTGTCGCAGGGCACCCCCGGACTCGGCTCCAAAGTCAAATGGGACCACTCGCTGGAGTGGAGCGTGGCTAACTTCAGCGTGCCTCCGTCCGGGGAAAATATCATCGAATTCGATCTGTCAAAGGCCGACGACGCCTTCATAGCGGGACACAATATAGACGGGCGCGTGCTGTTCCCCGCCACCGGGTACTTGACGCTCGTCTGGCGCACGATGGCCAAACTGAGCAACAGGAAGCTCGAGGACACCCCCATAGTGCTGGAGAACGTGCAGTTCAAGCGCGCGACCATCATGTCCAAGGACGGGCCGGTGCGTTTCCTCATCAACGTCCTCGAGGGCAGCGGACACTTCGACGTCTGCGAGGGCGGCTCCGTGGTCGTCACCGGCCTGGTCAGGCTGGCCGCGGACCCCGACAAGGAACGAATCCACGACGTCGAACCGGACAAGACCGAAGACTCCGAACCGCCACTGACCACGGAAGACATCTACAAAGAGCTGAGACTGAGAGGGTACAACTACGGCGGCATCTTCAAGGGGATCAAGGGGTCGGACGCGCGCGGTACTGTCGGGCAGCTCGCCTGGGAGGGAAACTGGATCTCGTACATCGACACCATGCTGCAGTTCGGCATCATCGGCGTCGACACCCGCGAGCTGTACCTGCCGACGCGGCTGCGGCGCGCGCTCATCGACCCCGCGGCGCTGGCGGAGCGGGCGCCGGGCGAGCCGGTCCGCGTCACCATGCGCAGAGACATCGACGTCATATCGGCGGCCGGAGTCGAACTGCGCGGCGTCAAGACCAGCCTGGCACCCAGGAGGGCCAACCCTCAGGCTCCTCCTAAACTGGAGAAATACGTGTTCCTGCCCTACGACAACACGTCTGTGGCCACCGAGGACACGTCGCGCTCCAAGAGAGACGCCATGACCGTTAGCTTGCAGATTGCTATGGAGAACATCGGAGCCCTGAAGCTGAAGGTGGCGGAGGCGGCCCTGTCCCGCGCACCCGAGGCGCTGCTGTCTCCTGCCGTGTCCGCGCTGCTGCACGCCGAGCCGCAGCAGCGCGCCGACGTGTCCGTGGGCGCGGGGCCCGACGCCGCCGCCTACAGTGCCGCGCTACAACAACACGACATCAAGGTGACGCCAAAAGACGGCCGCAGTGCTCCACTCGACGCAGACTGTCACCTCGTCGTAGCAGCTGACGTCATCGGGAGGCACGGCGCCCCCGTACTAAGGAACCTTATTGCGTCCCTGGCGCCCAACGGTCTACTGCTTCTAGAAGAACCACGCGGAGCTCTGAGCGAAAGAGCACCTCGCGACGTCATCGAACGCGAAGGGCTCGAGTTGATCTCGCGCCAAGTAGCCGCCCCGTGCGAGTACTTATTGTTGAGGCGCAAAGCTGAGCTCCCCTCGAGCCACGTGGTGGTGGACGTGTCCGACGACGTCAAGTTCGCCTGGGTGGAGGACCTGCGGGCGGCCCTGGGTCGTGCCGAAAACGAGAACATACGCGTGTATGCGGTCTCGCGGTCGGCGCACTGCGGAGCCCTGGGCCTGGCCACGTGCTTGCGCGCGGAGCCCGGGGGCGGCGCCCTGCGAGTGTACTATCTGCCTGGGGCCCGCGACCAGTTCGACGTGACGTCGCCCGCCTACTCGGGGCAGGTGCAGAAGGACTTGAGTGTGAACGTTCTGCGCTCCGGAGTGTGGGGGACCTACCGACACCTGCCCATCACCGACGCGGCCGATGCGCAGCTGCAGGTGGAGCACGCGTACGTGAATACCGTGACCCGGGGGGACCTGTCCTCGCTGTGCTGGATCGAGAGTCCACTGCGGTACGCGCAGGTGTTGGCACGGCCCCGGGGCACTGAGCTGTGCCGGGTGTACTGTGCGCCGCTCAACTTCCGGGACATCATGTTGGCCACCGGGAAACTGCCGCCGGACGCATTGCCCGGGAACTTGGCCGGACAGGAATGTATCCTTGGTCTCGAGTTCAGTGGACGCACCGAAAGTGGAAAACGCGTGATGGGCATGGTCGCCGCGCGCGGATTGGCCAGCACCGTGTTGGCAGACGAGGGCTTCCTCTGGGAGGTGCCCGAGGAGTGGTCGCTGGAGGAAGCGGCCACCGTGCCCGTGGCGTACGCCACCGCGTACTACGCGCTGGCGGTCCGCGGGGGCATGAGGCGCGGAGACGCCGTGCTGGTGCACGCGGGCTCGGGCGGGGTGGGGCAGGCGGCCATCGCCATCGCCCTGCACGCCGGCTGCGACGTGTTCACCACGGTCGGCACGCCGGACAAGCGGGCCTTCCTGCGCCAACGATTCCCGAAGCTGCCCGAAGAGAACATCGGCAACTCTCGGGACACGTCCTTCGAGCAGCTGGTGATGCGGCGCACGCGCGGCCGCGGGGTGGACCTGGTGCTCAACTCGCTGGCGGCGGACAAGCTGCACGCGTCGGTGCGCTGCCTCGCTGAAGGCGGACGGTTCTTGGAGATCGGTAAACTGGATCTCTCCGACGACACGCCCCTGGGCATGTCGATCTTCCTGAAGAACACAACATTCCACGGCATCCTGCTGGACGCGCTGTTCGATGCGGACAGCGAGCACCCGGACAAGCTGGCGGTGGTGCGCTGCGTGAGCGCGGGGCTGGCGGCCGGTGCTGTGCGCCCGCTGCCCGCCACCGTGTTCTCCGACACGCAGCTCGAGCCCGCATTCAGATACATGGCTGCAGGGAAGCACATCGGCAAGGTAGTGATCCGAGTCCGCGAGGAGGAGGCGGACGGAGCGCGGCCTGCCGCCCGGCTGATGGTGGCTGCGCCCCGCACCTACTTCCACCCGGCCAGGACGTACGTACTCGTCGGTGGACTGGGTGGGTTCGGACTGGAGTTGGCTCAGTGGCTGATCGTGCGCGGAGCCACTCGCCTCGTGCTCAACTCCCGCAGCGGAGTGCGCACGGGATACCAGGCCTGGTGCATACGCAGGTGGCGCTCGCGCGGCGTGCAGGTGTTGGTGTCGCGCACGGACAGCAGCACGGCGGAGGGCGCTCGCGCGCTGCTGCAGGCCGCCGCCGCGCTGGGGCCCGTGGGCGGACTGTTCAACCTGGCCGCCGTGCTCCGGGACGCCTTCCTCGACAAGCAGACGCCGGCCGACTTCCAGACCGTCGCCAAGCCCAAGATCGACGCTACCAAGGCCCTGGACGCGGCGAGCCGGGAGCTGGCCCCCGAGCTGGAGTACTTCGTGGTGTTCTCGTCGGTGTCGTGCGGGCGCGGCAACCCCGGACAGAGCAACTACGGCCTCGCCAACTCCGCCATGGAGCGGATCGTGGAGAGGAGACAGGCCGACGGGCTACCCGGCCTGGCCGTGCAGTGGGGCGCCATCGGTGAGGTCGGTCTGATCGTGGAGACGATGGGCGGCGACGACACGGAAGTGGGGGGCACGGTCCCGCAGCGCATCGCGTCCTGCATGGAGGCGCTGGGCGCGCTGCTGGCCCAGCCCCACGCCGTGGCCGCCAGCATGGTGCTCGCAGACAAGCGGCGCGCCCAGCAGGCGCCCAAGCAGGACCTGGTCCACGCTGTGGCCAACATACTGGGCATCAAGGATCCTAGCAAGGTGTCCGAATCCGCTAATCTAGCTGAACTCGGAATGGATTCTCTGATGGGCGCAGAGATCAAACAGACATTAGAGCGAGGGTACGACGTGGTCCTGGGAGTCCAGGAAATCCGAGCCCTGACTTTCGGTCGGCTGAAGGTGATGGCGGGGGTGGGGAGCGAGGGGGAGGGGGCCGGCGCCGACACCCCCCGCGGCCCCGCCGACCTGGTGCAGTTCGCGGCGCTCGGAGAACTCATGCCCAAGCAGACCCTCGTGAAACTGCCCAGCGCACAGCCGCCTTCTGCTGAGGCACCCCCTGTCTTCATGGTGCACCCTATAGAGGGCGTGGTGGACCTGCTCCGCGGCGTGGCGAGCGAGGTGGCGGGCGCGGTGTTCGGGCTGCAGTGCGCGGCCGGCGCGCCGCTGTCCTCCATGGCGGCGCTGGCGGAGCACTACGTCACGCACGTGCGCACCGTGCGCCCCCACCAGCCCTACCTGCTGCTGGGATACTCCTTCGGCGCCGCCGTGGCCTTCGAGATGGCGCTGCACCTCGAGGAGGCGGGCGCGGCCGTGCGGCTGGTGCTGGTGGACGGGTCGCCCGCGTACGTGGCCACGCACACGACGCGCGGGCGCACGCGCGGACAGTCGCGCGGCGGCTCGCGCGGACAACGCACGGGCCCCGCCGCCGCCGACGAGGCCGACGCGCTCGCATACTTCGCGCAGCTCTTCAGGGACATCGACGCCGCCAAGGTGTCAGCCGAACTAGAGCGTCTCCCGTCCTGGGAGAGTCGAGTGGAGCGCGTGACGTCACTGATCGACAGCGGCTCGCACAGCGCCGAGGCCCTGGCCGCGGCCGCCGCCTCGTTCTACAAGAAGCTGGTGATCGCGGACGCGTACAAGCCGACGCGGCAGCTGCGCGCGCCCGTCACGCTGTTCACGGCGCGCGACAACTACGTCACGCTCGGCGAGGACTACGGGCTCCGGGCTGTCTGCACGGGAGAGGTAAAGTGTCATCAGCTGGCGGGCACGCACCGCACGATACTGACGGGCGACGCGGCGCGCGACATCGCGTCGCATCTCTCCGCGCTGATCTAA
Protein
MPSVVNGTNGAGVDDDIVLTGLSGKLPESNNIEEFAEQLFAGVDLVTADSRRWVPGLHGLPERNGKLKDLAHFDATFFGVHAKQANLMDPQLRLLLELTHEAIIDAGFNPTELRGSRTGVYVGVSNSETEEMWTADPDKINGYALTGCCRAMFPNRISYTFDFCGPSYAVDTACSSSMFALAQAVTAMRAGHCDAAIVAGTNLCLKPANSLNFHRLSMLSPQGRCAAFDASGNGYVRSEAAVVVVLQRRANARRVYATVRGARMNTDGTKVQGITFPSGDMQRRLAKETFEEAGLRPQDVVYVEAHGTGTKVGDPQEVNAIAQLFCENRSDPLLLGSVKSNMGHSEPASGLCSIAKVVVAMENGVIPQNLHYKSPNLDIPALSDGRIKVVDRNLEWNGGLVAVNSFGFGGANAHIILESERVGKRVRTEYAIPRLVLASGRTEDAVSHLLEHAAKHAKDAELHGLLDAIHARNIPGHGYRGYGLLLNEMQTEIVETSSGDPRPIWYVFSGMGSQWAGMAKCLMRLPVFAASVHRSAAALRPHGIDLVHAITEAPDTAFDDVVISFVSIAAVQVAMVDVLRELQITPDGIVGHSVGEIGCAYADETLTGEQAVLAAYWRGRSIVDARLDAGAMAAVGLSWELCQRRCPPDVAPACHNASDNVTISGPVASIEKFVAELTAEGIFARRVNSSGIAFHSKYIAAAAPLLRKSLEKVIPYPKARSSKWVSSSLPRDQWGSELAKLSDVNYHVNNLLSPVRFADALKEIPERAIVVEIAPHALLQAVLKRAVPAPAAAHVALVRRDAADALLHLLGAVGKLYAAGAQPQPSRLYPAVRFPVSQGTPGLGSKVKWDHSLEWSVANFSVPPSGENIIEFDLSKADDAFIAGHNIDGRVLFPATGYLTLVWRTMAKLSNRKLEDTPIVLENVQFKRATIMSKDGPVRFLINVLEGSGHFDVCEGGSVVVTGLVRLAADPDKERIHDVEPDKTEDSEPPLTTEDIYKELRLRGYNYGGIFKGIKGSDARGTVGQLAWEGNWISYIDTMLQFGIIGVDTRELYLPTRLRRALIDPAALAERAPGEPVRVTMRRDIDVISAAGVELRGVKTSLAPRRANPQAPPKLEKYVFLPYDNTSVATEDTSRSKRDAMTVSLQIAMENIGALKLKVAEAALSRAPEALLSPAVSALLHAEPQQRADVSVGAGPDAAAYSAALQQHDIKVTPKDGRSAPLDADCHLVVAADVIGRHGAPVLRNLIASLAPNGLLLLEEPRGALSERAPRDVIEREGLELISRQVAAPCEYLLLRRKAELPSSHVVVDVSDDVKFAWVEDLRAALGRAENENIRVYAVSRSAHCGALGLATCLRAEPGGGALRVYYLPGARDQFDVTSPAYSGQVQKDLSVNVLRSGVWGTYRHLPITDAADAQLQVEHAYVNTVTRGDLSSLCWIESPLRYAQVLARPRGTELCRVYCAPLNFRDIMLATGKLPPDALPGNLAGQECILGLEFSGRTESGKRVMGMVAARGLASTVLADEGFLWEVPEEWSLEEAATVPVAYATAYYALAVRGGMRRGDAVLVHAGSGGVGQAAIAIALHAGCDVFTTVGTPDKRAFLRQRFPKLPEENIGNSRDTSFEQLVMRRTRGRGVDLVLNSLAADKLHASVRCLAEGGRFLEIGKLDLSDDTPLGMSIFLKNTTFHGILLDALFDADSEHPDKLAVVRCVSAGLAAGAVRPLPATVFSDTQLEPAFRYMAAGKHIGKVVIRVREEEADGARPAARLMVAAPRTYFHPARTYVLVGGLGGFGLELAQWLIVRGATRLVLNSRSGVRTGYQAWCIRRWRSRGVQVLVSRTDSSTAEGARALLQAAAALGPVGGLFNLAAVLRDAFLDKQTPADFQTVAKPKIDATKALDAASRELAPELEYFVVFSSVSCGRGNPGQSNYGLANSAMERIVERRQADGLPGLAVQWGAIGEVGLIVETMGGDDTEVGGTVPQRIASCMEALGALLAQPHAVAASMVLADKRRAQQAPKQDLVHAVANILGIKDPSKVSESANLAELGMDSLMGAEIKQTLERGYDVVLGVQEIRALTFGRLKVMAGVGSEGEGAGADTPRGPADLVQFAALGELMPKQTLVKLPSAQPPSAEAPPVFMVHPIEGVVDLLRGVASEVAGAVFGLQCAAGAPLSSMAALAEHYVTHVRTVRPHQPYLLLGYSFGAAVAFEMALHLEEAGAAVRLVLVDGSPAYVATHTTRGRTRGQSRGGSRGQRTGPAAADEADALAYFAQLFRDIDAAKVSAELERLPSWESRVERVTSLIDSGSHSAEALAAAAASFYKKLVIADAYKPTRQLRAPVTLFTARDNYVTLGEDYGLRAVCTGEVKCHQLAGTHRTILTGDAARDIASHLSALI
Summary
Uniprot
U5KGX9
A0A0F6Q2M8
A0A2H1WN88
A0A212FLS7
A0A194QPH6
A0A1B2M4Y5
+ More
V5GRN6 A0A1B6D462 A0A1W5YLL8 D6W6D2 A0A1B6DTB3 A0A1W4XJ56 A0A344X1T4 K7IM26 A0A1B6CY84 N6TYU8 A0A232F3H2 U4UFT3 E0W0Z3 J9K5E2 A0A084VMX1 A0A088AP68 A0A182PFR7 A0A182MD01 A0A182JUV3 A0A182J9V0 A0A182UXM1 A0A182WH69 V9IC11 A0A1S4EY19 V9IED1 A0A182GF80 Q17M16 A0A182R6I9 A0A1Q3FQU2 A0A2A3ESK1 W4VRN6 A0A1Q3FQM2 A0A1Q3FQI5 A0A182QS29 A0A182NF49 A0A0L0BS22 A0A1L8DZ80 A0A1L8DZ67 A0A1Q3FQT3 A0A310S575 A0A1L8DZ41 A0A182KUF7 A0A182I565 Q7PVV2 A0A182TT13 B0WEB3 A0A182WYK6 A0A182Y9S4 A0A182F1L9 A0A2M3YXF7 A0A1Q3FQC0 A0A2M4B865 W5J8N6 A0A1J1I5S9 B3MLJ5 A0A3B0JFS1 A0A1I8NSS5 A0A1W4UTP9 B4NXD6 Q29M08 B4G6R3 B3NAQ7 A0A1I8MI56 A0A195CLZ5 A0A151X557 A0A1A9WUD1 A0A026WXS0 T1PGU1 B4KI46 A0A0L7QN56 B7Z001 Q9VQL7 A0A154PP11 A0A0J9TEH0 A0A1A9XYR8 A0A0M9A0W5 B4NLM1 B4I2V4 A0A224X4N0 A0A195EVF8 A0A0A1XRS9 B4JBD0 A0A1B0FM30 A0A1A9Z5Z9 G9JWG5 H6WCY9 F4X257 A0A1A9UTY3 A0A0K8S3M9 A0A0A9XBY7 A0A0M3QT47 A0A195DQH9 A0A1D2N224 A0A158NAI9
V5GRN6 A0A1B6D462 A0A1W5YLL8 D6W6D2 A0A1B6DTB3 A0A1W4XJ56 A0A344X1T4 K7IM26 A0A1B6CY84 N6TYU8 A0A232F3H2 U4UFT3 E0W0Z3 J9K5E2 A0A084VMX1 A0A088AP68 A0A182PFR7 A0A182MD01 A0A182JUV3 A0A182J9V0 A0A182UXM1 A0A182WH69 V9IC11 A0A1S4EY19 V9IED1 A0A182GF80 Q17M16 A0A182R6I9 A0A1Q3FQU2 A0A2A3ESK1 W4VRN6 A0A1Q3FQM2 A0A1Q3FQI5 A0A182QS29 A0A182NF49 A0A0L0BS22 A0A1L8DZ80 A0A1L8DZ67 A0A1Q3FQT3 A0A310S575 A0A1L8DZ41 A0A182KUF7 A0A182I565 Q7PVV2 A0A182TT13 B0WEB3 A0A182WYK6 A0A182Y9S4 A0A182F1L9 A0A2M3YXF7 A0A1Q3FQC0 A0A2M4B865 W5J8N6 A0A1J1I5S9 B3MLJ5 A0A3B0JFS1 A0A1I8NSS5 A0A1W4UTP9 B4NXD6 Q29M08 B4G6R3 B3NAQ7 A0A1I8MI56 A0A195CLZ5 A0A151X557 A0A1A9WUD1 A0A026WXS0 T1PGU1 B4KI46 A0A0L7QN56 B7Z001 Q9VQL7 A0A154PP11 A0A0J9TEH0 A0A1A9XYR8 A0A0M9A0W5 B4NLM1 B4I2V4 A0A224X4N0 A0A195EVF8 A0A0A1XRS9 B4JBD0 A0A1B0FM30 A0A1A9Z5Z9 G9JWG5 H6WCY9 F4X257 A0A1A9UTY3 A0A0K8S3M9 A0A0A9XBY7 A0A0M3QT47 A0A195DQH9 A0A1D2N224 A0A158NAI9
Pubmed
24053512
22118469
26354079
18362917
19820115
20075255
+ More
23537049 28648823 20566863 24438588 26483478 17510324 26108605 20966253 12364791 25244985 20920257 23761445 17994087 17550304 15632085 25315136 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 25830018 23325667 21719571 25401762 26823975 27289101 21347285
23537049 28648823 20566863 24438588 26483478 17510324 26108605 20966253 12364791 25244985 20920257 23761445 17994087 17550304 15632085 25315136 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 25830018 23325667 21719571 25401762 26823975 27289101 21347285
EMBL
JX989151
AGR49310.1
KP237908
AKD01761.1
ODYU01009790
SOQ54446.1
+ More
AGBW02007696 OWR54692.1 KQ461185 KPJ07407.1 KX467777 AOA60273.1 GALX01004174 JAB64292.1 GEDC01016862 GEDC01012243 JAS20436.1 JAS25055.1 KX147671 ARI45075.1 KQ971319 EFA11365.1 GEDC01031722 GEDC01008387 JAS05576.1 JAS28911.1 MF817713 AXE71619.1 GEDC01031101 GEDC01028079 GEDC01018856 GEDC01013285 JAS06197.1 JAS09219.1 JAS18442.1 JAS24013.1 APGK01049539 APGK01049540 KB741156 ENN73526.1 NNAY01001061 OXU25291.1 KB632169 ERL89446.1 DS235863 EEB19298.1 ABLF02040018 ATLV01014632 KE524975 KFB39315.1 AXCM01004007 JR039354 AEY58653.1 JR039355 AEY58654.1 JXUM01059545 JXUM01059546 JXUM01059547 CH477209 EAT47728.1 GFDL01005120 JAV29925.1 KZ288193 PBC34021.1 GANO01001242 JAB58629.1 GFDL01005223 JAV29822.1 GFDL01005164 JAV29881.1 AXCN02000829 JRES01001454 KNC22821.1 GFDF01002357 JAV11727.1 GFDF01002358 JAV11726.1 GFDL01005130 JAV29915.1 KQ770827 OAD52584.1 GFDF01002356 JAV11728.1 APCN01003789 AAAB01008984 EAA15087.4 DS231907 EDS45473.1 GGFM01000190 MBW20941.1 GFDL01005234 JAV29811.1 GGFJ01000040 MBW49181.1 ADMH02001861 ETN60822.1 CVRI01000038 CRK93737.1 CH902620 EDV31744.2 OUUW01000004 SPP79543.1 CM000157 EDW87493.2 CH379060 EAL33887.3 CH479180 EDW29177.1 CH954177 EDV57580.2 KQ977642 KYN01094.1 KQ982519 KYQ55543.1 KK107077 QOIP01000010 EZA60541.1 RLU17791.1 KA647365 AFP61994.1 CH933807 EDW12339.1 KQ414860 KOC60060.1 AE014134 ACL82985.2 AAF51148.1 KQ435007 KZC13477.1 CM002910 KMY87820.1 KQ435796 KOX73549.1 CH964274 EDW85260.2 CH480820 EDW54099.1 GFTR01008959 JAW07467.1 KQ981954 KYN32228.1 GBXI01000610 JAD13682.1 CH916368 EDW02935.1 CCAG010019592 JN982132 AEW43642.1 JQ177158 AEY83835.1 GL888565 EGI59473.1 GBRD01017936 JAG47891.1 GBHO01026473 GBRD01017935 GDHC01015482 JAG17131.1 JAG47892.1 JAQ03147.1 CP012523 ALC38283.1 KQ980612 KYN15123.1 LJIJ01000286 ODM99347.1 ADTU01010333 ADTU01010334
AGBW02007696 OWR54692.1 KQ461185 KPJ07407.1 KX467777 AOA60273.1 GALX01004174 JAB64292.1 GEDC01016862 GEDC01012243 JAS20436.1 JAS25055.1 KX147671 ARI45075.1 KQ971319 EFA11365.1 GEDC01031722 GEDC01008387 JAS05576.1 JAS28911.1 MF817713 AXE71619.1 GEDC01031101 GEDC01028079 GEDC01018856 GEDC01013285 JAS06197.1 JAS09219.1 JAS18442.1 JAS24013.1 APGK01049539 APGK01049540 KB741156 ENN73526.1 NNAY01001061 OXU25291.1 KB632169 ERL89446.1 DS235863 EEB19298.1 ABLF02040018 ATLV01014632 KE524975 KFB39315.1 AXCM01004007 JR039354 AEY58653.1 JR039355 AEY58654.1 JXUM01059545 JXUM01059546 JXUM01059547 CH477209 EAT47728.1 GFDL01005120 JAV29925.1 KZ288193 PBC34021.1 GANO01001242 JAB58629.1 GFDL01005223 JAV29822.1 GFDL01005164 JAV29881.1 AXCN02000829 JRES01001454 KNC22821.1 GFDF01002357 JAV11727.1 GFDF01002358 JAV11726.1 GFDL01005130 JAV29915.1 KQ770827 OAD52584.1 GFDF01002356 JAV11728.1 APCN01003789 AAAB01008984 EAA15087.4 DS231907 EDS45473.1 GGFM01000190 MBW20941.1 GFDL01005234 JAV29811.1 GGFJ01000040 MBW49181.1 ADMH02001861 ETN60822.1 CVRI01000038 CRK93737.1 CH902620 EDV31744.2 OUUW01000004 SPP79543.1 CM000157 EDW87493.2 CH379060 EAL33887.3 CH479180 EDW29177.1 CH954177 EDV57580.2 KQ977642 KYN01094.1 KQ982519 KYQ55543.1 KK107077 QOIP01000010 EZA60541.1 RLU17791.1 KA647365 AFP61994.1 CH933807 EDW12339.1 KQ414860 KOC60060.1 AE014134 ACL82985.2 AAF51148.1 KQ435007 KZC13477.1 CM002910 KMY87820.1 KQ435796 KOX73549.1 CH964274 EDW85260.2 CH480820 EDW54099.1 GFTR01008959 JAW07467.1 KQ981954 KYN32228.1 GBXI01000610 JAD13682.1 CH916368 EDW02935.1 CCAG010019592 JN982132 AEW43642.1 JQ177158 AEY83835.1 GL888565 EGI59473.1 GBRD01017936 JAG47891.1 GBHO01026473 GBRD01017935 GDHC01015482 JAG17131.1 JAG47892.1 JAQ03147.1 CP012523 ALC38283.1 KQ980612 KYN15123.1 LJIJ01000286 ODM99347.1 ADTU01010333 ADTU01010334
Proteomes
UP000007151
UP000053240
UP000007266
UP000192223
UP000002358
UP000019118
+ More
UP000215335 UP000030742 UP000009046 UP000007819 UP000030765 UP000005203 UP000075885 UP000075883 UP000075881 UP000075880 UP000075903 UP000075920 UP000069940 UP000008820 UP000075900 UP000242457 UP000075886 UP000075884 UP000037069 UP000075882 UP000075840 UP000007062 UP000075902 UP000002320 UP000076407 UP000076408 UP000069272 UP000000673 UP000183832 UP000007801 UP000268350 UP000095300 UP000192221 UP000002282 UP000001819 UP000008744 UP000008711 UP000095301 UP000078542 UP000075809 UP000091820 UP000053097 UP000279307 UP000009192 UP000053825 UP000000803 UP000076502 UP000092443 UP000053105 UP000007798 UP000001292 UP000078541 UP000001070 UP000092444 UP000092445 UP000007755 UP000078200 UP000092553 UP000078492 UP000094527 UP000005205
UP000215335 UP000030742 UP000009046 UP000007819 UP000030765 UP000005203 UP000075885 UP000075883 UP000075881 UP000075880 UP000075903 UP000075920 UP000069940 UP000008820 UP000075900 UP000242457 UP000075886 UP000075884 UP000037069 UP000075882 UP000075840 UP000007062 UP000075902 UP000002320 UP000076407 UP000076408 UP000069272 UP000000673 UP000183832 UP000007801 UP000268350 UP000095300 UP000192221 UP000002282 UP000001819 UP000008744 UP000008711 UP000095301 UP000078542 UP000075809 UP000091820 UP000053097 UP000279307 UP000009192 UP000053825 UP000000803 UP000076502 UP000092443 UP000053105 UP000007798 UP000001292 UP000078541 UP000001070 UP000092444 UP000092445 UP000007755 UP000078200 UP000092553 UP000078492 UP000094527 UP000005205
Pfam
Interpro
IPR016035
Acyl_Trfase/lysoPLipase
+ More
IPR023102 Fatty_acid_synthase_dom_2
IPR014031 Ketoacyl_synth_C
IPR036736 ACP-like_sf
IPR016036 Malonyl_transacylase_ACP-bd
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR001031 Thioesterase
IPR018201 Ketoacyl_synth_AS
IPR011032 GroES-like_sf
IPR020806 PKS_PP-bd
IPR042104 PKS_dehydratase_sf
IPR009081 PP-bd_ACP
IPR032821 KAsynt_C_assoc
IPR020801 PKS_acyl_transferase
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR013968 PKS_KR
IPR016039 Thiolase-like
IPR029058 AB_hydrolase
IPR001227 Ac_transferase_dom_sf
IPR020807 PKS_dehydratase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR013149 ADH_C
IPR023102 Fatty_acid_synthase_dom_2
IPR014031 Ketoacyl_synth_C
IPR036736 ACP-like_sf
IPR016036 Malonyl_transacylase_ACP-bd
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR001031 Thioesterase
IPR018201 Ketoacyl_synth_AS
IPR011032 GroES-like_sf
IPR020806 PKS_PP-bd
IPR042104 PKS_dehydratase_sf
IPR009081 PP-bd_ACP
IPR032821 KAsynt_C_assoc
IPR020801 PKS_acyl_transferase
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR013968 PKS_KR
IPR016039 Thiolase-like
IPR029058 AB_hydrolase
IPR001227 Ac_transferase_dom_sf
IPR020807 PKS_dehydratase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR013149 ADH_C
SUPFAM
ProteinModelPortal
U5KGX9
A0A0F6Q2M8
A0A2H1WN88
A0A212FLS7
A0A194QPH6
A0A1B2M4Y5
+ More
V5GRN6 A0A1B6D462 A0A1W5YLL8 D6W6D2 A0A1B6DTB3 A0A1W4XJ56 A0A344X1T4 K7IM26 A0A1B6CY84 N6TYU8 A0A232F3H2 U4UFT3 E0W0Z3 J9K5E2 A0A084VMX1 A0A088AP68 A0A182PFR7 A0A182MD01 A0A182JUV3 A0A182J9V0 A0A182UXM1 A0A182WH69 V9IC11 A0A1S4EY19 V9IED1 A0A182GF80 Q17M16 A0A182R6I9 A0A1Q3FQU2 A0A2A3ESK1 W4VRN6 A0A1Q3FQM2 A0A1Q3FQI5 A0A182QS29 A0A182NF49 A0A0L0BS22 A0A1L8DZ80 A0A1L8DZ67 A0A1Q3FQT3 A0A310S575 A0A1L8DZ41 A0A182KUF7 A0A182I565 Q7PVV2 A0A182TT13 B0WEB3 A0A182WYK6 A0A182Y9S4 A0A182F1L9 A0A2M3YXF7 A0A1Q3FQC0 A0A2M4B865 W5J8N6 A0A1J1I5S9 B3MLJ5 A0A3B0JFS1 A0A1I8NSS5 A0A1W4UTP9 B4NXD6 Q29M08 B4G6R3 B3NAQ7 A0A1I8MI56 A0A195CLZ5 A0A151X557 A0A1A9WUD1 A0A026WXS0 T1PGU1 B4KI46 A0A0L7QN56 B7Z001 Q9VQL7 A0A154PP11 A0A0J9TEH0 A0A1A9XYR8 A0A0M9A0W5 B4NLM1 B4I2V4 A0A224X4N0 A0A195EVF8 A0A0A1XRS9 B4JBD0 A0A1B0FM30 A0A1A9Z5Z9 G9JWG5 H6WCY9 F4X257 A0A1A9UTY3 A0A0K8S3M9 A0A0A9XBY7 A0A0M3QT47 A0A195DQH9 A0A1D2N224 A0A158NAI9
V5GRN6 A0A1B6D462 A0A1W5YLL8 D6W6D2 A0A1B6DTB3 A0A1W4XJ56 A0A344X1T4 K7IM26 A0A1B6CY84 N6TYU8 A0A232F3H2 U4UFT3 E0W0Z3 J9K5E2 A0A084VMX1 A0A088AP68 A0A182PFR7 A0A182MD01 A0A182JUV3 A0A182J9V0 A0A182UXM1 A0A182WH69 V9IC11 A0A1S4EY19 V9IED1 A0A182GF80 Q17M16 A0A182R6I9 A0A1Q3FQU2 A0A2A3ESK1 W4VRN6 A0A1Q3FQM2 A0A1Q3FQI5 A0A182QS29 A0A182NF49 A0A0L0BS22 A0A1L8DZ80 A0A1L8DZ67 A0A1Q3FQT3 A0A310S575 A0A1L8DZ41 A0A182KUF7 A0A182I565 Q7PVV2 A0A182TT13 B0WEB3 A0A182WYK6 A0A182Y9S4 A0A182F1L9 A0A2M3YXF7 A0A1Q3FQC0 A0A2M4B865 W5J8N6 A0A1J1I5S9 B3MLJ5 A0A3B0JFS1 A0A1I8NSS5 A0A1W4UTP9 B4NXD6 Q29M08 B4G6R3 B3NAQ7 A0A1I8MI56 A0A195CLZ5 A0A151X557 A0A1A9WUD1 A0A026WXS0 T1PGU1 B4KI46 A0A0L7QN56 B7Z001 Q9VQL7 A0A154PP11 A0A0J9TEH0 A0A1A9XYR8 A0A0M9A0W5 B4NLM1 B4I2V4 A0A224X4N0 A0A195EVF8 A0A0A1XRS9 B4JBD0 A0A1B0FM30 A0A1A9Z5Z9 G9JWG5 H6WCY9 F4X257 A0A1A9UTY3 A0A0K8S3M9 A0A0A9XBY7 A0A0M3QT47 A0A195DQH9 A0A1D2N224 A0A158NAI9
PDB
6FN6
E-value=0,
Score=2793
Ontologies
GO
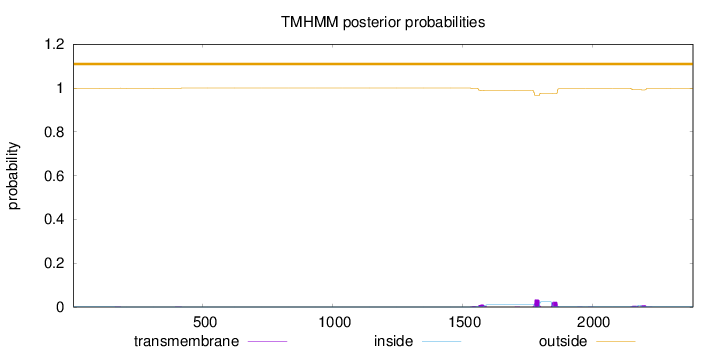
Topology
Length:
2389
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.86128
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00070
outside
1 - 2389
Population Genetic Test Statistics
Pi
19.682466
Theta
17.775713
Tajima's D
-1.265867
CLR
2.029335
CSRT
0.0893455327233638
Interpretation
Uncertain
