Gene
KWMTBOMO06931
Pre Gene Modal
BGIBMGA004656
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_myosin_IA_heavy_chain_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.246 Nuclear Reliability : 1.529 PlasmaMembrane Reliability : 1.089
Sequence
CDS
ATGGGCAGGTCTCGTTTTGTCGGTGACCTATCGAGGTCGTTCAAACGCTCAGGAAATACCATTTCTAGTCGAAAAGTAGGTGTCGCTCGTGCTGTGCTGCGGGGTGGGGGCAGGGCGGGAGGTGAGGGGGAGCGTTGCGCGGCCGGGCCGCGGGGCGGGGCGGGCGGAGGCTTTAGTAAACGATCGTGTGCGAGCGCGAGAGGACGCCGCCGAGTTAGAAGCGGCGCGGCCGCGGAGTGTGATGCGAGCGCGCCGCCCGCCATGGCCACGCTCGGCCTCTCCAAGGTGTTCATCCTCGACAAGTACTTCACCGAGCTGCAGAAGTTCTGGGAGACTGAGAAGAAGCTGCAAGACGCGTCGTCATCGAACGAGGCGGTGCACCTGCAACGACGGCTGCTGAGTCTGAGCTCGGAGCTGGTGACGCTTCGCAACCACTTGCACGTGGGAGCGGCTGGAGGCACGGCTGCCGGTGCGGCCCCCACTGCCGGCACTAGCACCGGACAACCCGCCGTCCCGCCGAGAGCACCGCACTTGTTACCACCGCCGCCGCACGCCGCGCAACCGCCCGCTCCACCGCTTCACCCCCTGCCGCCTCCTCCTGAAACGCGATGGCGGAACGTTCCGGCCGCCCCACCACAAGCGAGCGCGGGCCCTACTCCCACGAGTGGGACGAGCGGTCCGAGCGGAGTGAGCGGTCTAAGCGCAAGTGACGTGGAGGACCTCATACATCTAAGAGGACCGCTCACAGAAGACGCGGTCGTTAGAGCGTTGCAGGCGAGATTCTATCACAATAAGTTTTATACGATGATCGGACCAATCCTCATCGCGGTAAATCCATACACGGACACCTGTAACGCATTAACGTTGACTGCGGCCCGCGCCCACCGGCCCGAGCTAGCCCGACTGGTTCACGACGCCGTGAGACACCAGGCGGACTCAGGGTACCCTCAGGCGATCATTCTCTCAGGGGTATCTGGCTCAGGGAAGACGTACGCGTCAATGGTGTTGTTGAGGAGGCTCTTCGACGTGGCCGGTGGAGGCCCCGAAACGGATGCGTTCAAGCATCTCGCGGCGGCTTTTACTGTGCTGAGGGCTTTGGGCACGGCCGCCACACCGGCTAATGCGCATTCGTCCCGTATAGGACATTTCATCGAGGTCCAAGTAACCGACGGAGCCTTGTATCGTACAAAGATCCACTGTTACTTCCTCGACCAGACCAGAGTGGTCCGCCCTCCAGCTGGCGAACGAAACTATCACATCTTCTACCAGATGCTGGCCGGACTCACACCCGACGAGCGATCGCAACTTCATCTTGAAGGTTATTCCGCTCAGGACTTAAGGTATCTAGCATCCCCCCATCACCGTAGAGCAGAGCCAGAAGATGCTGCAAGATTCCATGCGTGGAAAACCTGTCTCGTTATTCTCGGTATACCGTTCCTTGACGTAGTACGAGTTTTAGCGGCTGTCCTATTACTTGGCAACGTTCAGTTCGCCGAGAGCGCTGAGGGTGGGGCCGAGCCAGCCGGAGAAGCAGAGCTAGCAGCAGCCGCGGCTCTGCTAGGAGTTGCAGCGCCGGCATTGTTACGAGGCTTGGCAATGCGGGCTCCACCTCGGACTTCATCTGCGCGATCAGTGCGCGTACCAGCCACCCCAGCGGCGGCTTCATCAGCGCGTGATGCACTGGCCAAAGCATTGTATTGCCGAACAGTGGCGACTATAGTGCGTAGAGCCAATTCGTTGAAACGCTTAGGATCTACTTTAGGCACGCTTTCGTCGGATTCAACTGAATCGGTACACGCGCAAGAAGCACGTCGAGCGTCATCAGCAGGCGGGTCTCGAGCGGGCGCTCGTTCAATGGCTGCACTTAACGACGCCGTACGACACGCCACTGATGGGTTCGTGGGGATATTGGACATGTTCGGTTTCGAGGACGCAGCCCCGAGTCGACTGGAGCACCTGTGCGTTAATCTGTGCGCAGAAACAATGCAACACTTTTATAATACTCACGTGTTCAAATCCGCCGCTGAATCTTGTCGCGAGGAAGGTGTGGCGTGTCCGCTCGAAGTGGAATACGTCGACAATGTGCCGTGTATTGATTTGGTGTCATCCTTGCGAGGCGGACTGCTTGCTGCACTGGACGCAGAATGTGCAGCCCGTGGTACTCCAGAACAATACGTAGTGCGCATTAAAACAGCACACAGAGGTCATCCGCGACTGGCGGAGGCTCGTCCACCGCACCCTCGAAGGTTTGCAGTGCGGCATTATGCTGGTGAAGTGGCATACGACACGGTGGACTTCTTGGAAGCTAATCGCGACGCCGTCCCTGACGACCTCCTTGCCGCCTTCGAAACTGGAACCTGTGAATTTGGTTTTGCTACACATCTTTTCGGGGCTGAACTTAAAGCCATGGCAGCACAAGGCGGGGCAGCGAATTCTGGGGGTGGATTCTTCCGTGCGTCCCCCACAGTTGCTGCAGAAGCAGGCTCGACACTCACACAAGACTTCCACACGCGTCTCGACAACTTGCTCCGTACGCTCGTGCACGCTCGCCCGCACTTCGTGCGCTGTTTGCGGGCTAACTCTACGGAAACACCAATGCTCTTTGAAAGGACGACTGTAGCGCGACAGGTCCGTGCACTACAAATATTAGAAACGACGCAGCTAATGGCCAGTGGGTACCCGCACCGGATGCGTTTCCGTGCGTTCAGCGGACGGTACCGGGCACTGTGCCGCGGCGCGGGGGGCGGGGCGAGTGCGGGGGCTGCGGGGAGCGTGGCGGGCGCGGAGGGGGAGGCGTGCGGGCGGGTACTGAGGGCAGTGCAGGCTGCGGCCGCTCCACCAGCCCCCGCCTCCCCTGCCGCAGTACGGTGGGCGCTTGGAAAGAGACACGTCTTCCTCAGTGAGGGGATGCGGCAGGTGTTAGAGCGGATGCGACGTGGTCGTCGTGAAGCGGCCGCTGCGAGTATACAGCGTGCGTGGCGGCGACACCGCGCCCGCCCCCGCCCCGCGCCGCGCCCCCGCCCCGCGCCCATCGCGCGCACCCCGCCTCCCGACCCCGCCGACAAGTGCGACCCCGCGCTCGTTAAGCGAACGTGCTCGCTCTTTGGACTCGACCTCGAGCGTCCCCCACCGCTGCCCCCGTCCCGCGCGTACACCGTGTCGGGGGGCGTGAAGCTGAGCTACCCGCAGCAGCGCGCCGTGAGTGCCGAGTGGAGCGACGAGGGCGGCGCGCGCCTGCGCCCCGGCGACACCGTGCTCGTGGTCGGCGCCGCCCGCCGGGGACACCTCGCTGTACAGGTCCGATACAACACCTTTTTTTTATTGTCGTATTTTACTTGA
Protein
MGRSRFVGDLSRSFKRSGNTISSRKVGVARAVLRGGGRAGGEGERCAAGPRGGAGGGFSKRSCASARGRRRVRSGAAAECDASAPPAMATLGLSKVFILDKYFTELQKFWETEKKLQDASSSNEAVHLQRRLLSLSSELVTLRNHLHVGAAGGTAAGAAPTAGTSTGQPAVPPRAPHLLPPPPHAAQPPAPPLHPLPPPPETRWRNVPAAPPQASAGPTPTSGTSGPSGVSGLSASDVEDLIHLRGPLTEDAVVRALQARFYHNKFYTMIGPILIAVNPYTDTCNALTLTAARAHRPELARLVHDAVRHQADSGYPQAIILSGVSGSGKTYASMVLLRRLFDVAGGGPETDAFKHLAAAFTVLRALGTAATPANAHSSRIGHFIEVQVTDGALYRTKIHCYFLDQTRVVRPPAGERNYHIFYQMLAGLTPDERSQLHLEGYSAQDLRYLASPHHRRAEPEDAARFHAWKTCLVILGIPFLDVVRVLAAVLLLGNVQFAESAEGGAEPAGEAELAAAAALLGVAAPALLRGLAMRAPPRTSSARSVRVPATPAAASSARDALAKALYCRTVATIVRRANSLKRLGSTLGTLSSDSTESVHAQEARRASSAGGSRAGARSMAALNDAVRHATDGFVGILDMFGFEDAAPSRLEHLCVNLCAETMQHFYNTHVFKSAAESCREEGVACPLEVEYVDNVPCIDLVSSLRGGLLAALDAECAARGTPEQYVVRIKTAHRGHPRLAEARPPHPRRFAVRHYAGEVAYDTVDFLEANRDAVPDDLLAAFETGTCEFGFATHLFGAELKAMAAQGGAANSGGGFFRASPTVAAEAGSTLTQDFHTRLDNLLRTLVHARPHFVRCLRANSTETPMLFERTTVARQVRALQILETTQLMASGYPHRMRFRAFSGRYRALCRGAGGGASAGAAGSVAGAEGEACGRVLRAVQAAAAPPAPASPAAVRWALGKRHVFLSEGMRQVLERMRRGRREAAAASIQRAWRRHRARPRPAPRPRPAPIARTPPPDPADKCDPALVKRTCSLFGLDLERPPPLPPSRAYTVSGGVKLSYPQQRAVSAEWSDEGGARLRPGDTVLVVGAARRGHLAVQVRYNTFFLLSYFT
Summary
Uniprot
ProteinModelPortal
PDB
4ZLK
E-value=8.23603e-35,
Score=373
Ontologies
GO
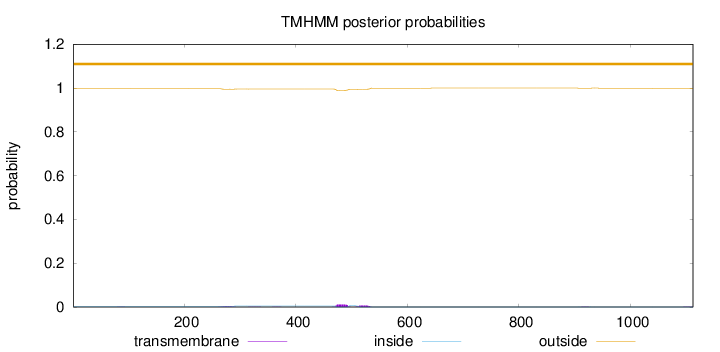
Topology
Length:
1112
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.468900000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00335
outside
1 - 1112
Population Genetic Test Statistics
Pi
23.469533
Theta
22.302546
Tajima's D
0.166706
CLR
1.565115
CSRT
0.418329083545823
Interpretation
Uncertain
