Gene
KWMTBOMO06929 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004657
Annotation
proteasome_subunit_alpha_type_6-A_[Bombyx_mori]
Full name
Proteasome subunit alpha type
+ More
Proteasome endopeptidase complex
Proteasome endopeptidase complex
Location in the cell
Mitochondrial Reliability : 1.284
Sequence
CDS
ATGGCTCGAGAAAGCAGTGCCGGATTCGACAGACATATTACTATCTTTTCACCTGAGGGAAGACTGTATCAAGTTGAATATGCATTGAAAGCTATAAACCAGGGTGGGTTGACATCTGTGGCATTACGCGGTACAGATGCAGCTGTAGTGGCAGCTCAACGCAAGGTCCCAGACAGACTACTTGATCCTGTCTCTGTTACTCATCTATTTGCTCTTACTGATAGAATTGGATGTGTCATGACCGGGATGATTGCTGATAGCCGGTCTCAAGTACAACGAGCTCGCTACGAGGCTGCCAACTGGCAGTACAAGTATGGGTCTGAGATACCTGTGCATATCTTATGCAGACGCATAGCTGATATCTCTCAAGTTTACACCCAAAATGCAGAGATGAGACCTCTGGGTTGCAGTATGATGCTGATAGCGTACGACGATGAAGCCGGGCCCTGTGTGTACAAAACAGACCCCTCCGGCTACTACTGCTCCTATAAAGCTGTAGCTGCTGGTGCTAAGACAGTTGATGGAAACGCTTATCTCGAAAAGAAACTAAAAAAACGCTCGGACCTCAGCACCGATGATGCGATTCAGCTTGCCATCTCTTGTCTGTCTCAAGTGTTGTCTGTCGACTTCAAGTCCTCTGAGATTGAGATTGGTGTTGTCAGCAAAGATAATCCTAAGTTTCGGGTGTTGACTGAAACTGAAATCGACAGACATCTCACGGCAATCGCTGAAAAGGATTAA
Protein
MARESSAGFDRHITIFSPEGRLYQVEYALKAINQGGLTSVALRGTDAAVVAAQRKVPDRLLDPVSVTHLFALTDRIGCVMTGMIADSRSQVQRARYEAANWQYKYGSEIPVHILCRRIADISQVYTQNAEMRPLGCSMMLIAYDDEAGPCVYKTDPSGYYCSYKAVAAGAKTVDGNAYLEKKLKKRSDLSTDDAIQLAISCLSQVLSVDFKSSEIEIGVVSKDNPKFRVLTETEIDRHLTAIAEKD
Summary
Description
The proteasome is a multicatalytic proteinase complex which is characterized by its ability to cleave peptides with Arg, Phe, Tyr, Leu, and Glu adjacent to the leaving group at neutral or slightly basic pH.
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Subunit
The 20S proteasome core is composed of 28 subunits that are arranged in four stacked rings, resulting in a barrel-shaped structure. The two end rings are each formed by seven alpha subunits, and the two central rings are each formed by seven beta subunits.
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
Similarity
Belongs to the peptidase T1A family.
Uniprot
Q1HPR1
H9J567
A0A2W1BTA5
A0A2A4JY31
A0A2H1W7W9
A0A1E1WKL4
+ More
A0A212FLT9 A0A3S2PH76 S4PTB5 A0A194QV38 I4DMJ9 I4DKX7 A0A2J7QYC4 A0A151I7N5 E2C5K7 A0A151JNM4 F4WKP2 A0A158NJ80 A0A195BJP3 A0A151WEA7 A0A1B6J933 A0A1B6H343 E2AJ67 K7ISL4 A0A026WXD0 A0A0J7N1R1 A0A1B6ECE7 A0A151JV70 R4UVY8 E9J1R8 A0A2A3E585 A0A1Q3FFG7 B0W3G3 A0A067R5N5 Q16UH5 E0VLY3 A0A182G942 A0A023EL07 R4G8P0 E2J7E5 A2IAA3 A0A1B0CBQ7 A0A224XVB3 U5EX64 A0A069DQ64 A0A0V0G3S8 A0A182PK35 A0A1L8DSK5 A0A0L7RBS3 A0A084W942 A0A023FCW9 T1E8H5 A0A2M3Z7L5 A0A2M4AF32 A0A2M4BYA8 A0A182FNY3 A0A0P4VNA4 A0A182J2G1 A0A182VAX2 W5J336 A0A182U7E3 A0A182XH14 A0A182KMV9 Q7Q2B0 A0A182I8D9 A0A182N6S9 A0A182XXL2 A0A0K8TNR5 A0A182M649 A0A182KFJ6 C3XZ66 A0A0P5YRK1 A0A1S3K3R9 A0A164PR99 A0A1D2NKE6 A0A232F000 A0A182Q3Z1 A0A182RY65 A0A1J1ISY9 A0A182VQG5 A0A0N8ECU1 A0A0P5AIU9 A0A0P4YNB4 A0A0P5MC31 A0A0P5BCN4 A0A0P4ZF56 E9HAP5 A0A0P5IEI8 A0A0P5ASX6 A0A0N8AJF2 A0A0P4ZB74 A0A2R7X246 A0A0P4YTZ2 A0A0N8BH29 V9L6Y5 A0A0P5WZX1 T1JGS3 R7UBE7 A0A131Z275 K4GK00
A0A212FLT9 A0A3S2PH76 S4PTB5 A0A194QV38 I4DMJ9 I4DKX7 A0A2J7QYC4 A0A151I7N5 E2C5K7 A0A151JNM4 F4WKP2 A0A158NJ80 A0A195BJP3 A0A151WEA7 A0A1B6J933 A0A1B6H343 E2AJ67 K7ISL4 A0A026WXD0 A0A0J7N1R1 A0A1B6ECE7 A0A151JV70 R4UVY8 E9J1R8 A0A2A3E585 A0A1Q3FFG7 B0W3G3 A0A067R5N5 Q16UH5 E0VLY3 A0A182G942 A0A023EL07 R4G8P0 E2J7E5 A2IAA3 A0A1B0CBQ7 A0A224XVB3 U5EX64 A0A069DQ64 A0A0V0G3S8 A0A182PK35 A0A1L8DSK5 A0A0L7RBS3 A0A084W942 A0A023FCW9 T1E8H5 A0A2M3Z7L5 A0A2M4AF32 A0A2M4BYA8 A0A182FNY3 A0A0P4VNA4 A0A182J2G1 A0A182VAX2 W5J336 A0A182U7E3 A0A182XH14 A0A182KMV9 Q7Q2B0 A0A182I8D9 A0A182N6S9 A0A182XXL2 A0A0K8TNR5 A0A182M649 A0A182KFJ6 C3XZ66 A0A0P5YRK1 A0A1S3K3R9 A0A164PR99 A0A1D2NKE6 A0A232F000 A0A182Q3Z1 A0A182RY65 A0A1J1ISY9 A0A182VQG5 A0A0N8ECU1 A0A0P5AIU9 A0A0P4YNB4 A0A0P5MC31 A0A0P5BCN4 A0A0P4ZF56 E9HAP5 A0A0P5IEI8 A0A0P5ASX6 A0A0N8AJF2 A0A0P4ZB74 A0A2R7X246 A0A0P4YTZ2 A0A0N8BH29 V9L6Y5 A0A0P5WZX1 T1JGS3 R7UBE7 A0A131Z275 K4GK00
EC Number
3.4.25.1
Pubmed
19121390
28756777
22118469
23622113
26354079
22651552
+ More
20798317 21719571 21347285 20075255 24508170 30249741 21282665 24845553 17510324 20566863 26483478 24945155 17437641 26334808 24438588 25474469 27129103 20920257 23761445 20966253 12364791 14747013 17210077 25244985 26369729 18563158 27289101 28648823 21292972 24402279 23254933 26830274 23056606
20798317 21719571 21347285 20075255 24508170 30249741 21282665 24845553 17510324 20566863 26483478 24945155 17437641 26334808 24438588 25474469 27129103 20920257 23761445 20966253 12364791 14747013 17210077 25244985 26369729 18563158 27289101 28648823 21292972 24402279 23254933 26830274 23056606
EMBL
DQ443341
ABF51430.1
BABH01039392
KZ149906
PZC78302.1
NWSH01000369
+ More
PCG76925.1 PCG76932.1 ODYU01006893 SOQ49147.1 GDQN01003648 JAT87406.1 AGBW02007696 OWR54697.1 RSAL01000034 RVE51372.1 GAIX01012328 JAA80232.1 KQ461185 KPJ07421.1 AK402517 BAM19139.1 AK401945 KQ459252 BAM18567.1 KPJ02194.1 NEVH01009101 PNF33583.1 KQ978411 KYM94100.1 GL452770 EFN76820.1 KQ978858 KYN27927.1 GL888206 EGI65134.1 ADTU01017554 KQ976453 KYM85393.1 KQ983247 KYQ46160.1 GECU01011980 JAS95726.1 GECZ01000676 JAS69093.1 GL439967 EFN66507.1 KK107078 QOIP01000011 EZA60401.1 RLU16699.1 LBMM01011888 KMQ86570.1 GEDC01001697 JAS35601.1 KQ981703 KYN37439.1 KC571887 AGM32386.1 GL767674 EFZ13292.1 KZ288363 PBC26923.1 GFDL01008793 JAV26252.1 DS231831 EDS31306.1 KK852869 KDR14657.1 CH477622 EAT38162.1 DS235283 EEB14389.1 JXUM01049009 KQ561586 KXJ78094.1 GAPW01003710 JAC09888.1 ACPB03006555 GAHY01000637 JAA76873.1 HP429363 ADN29863.1 EF179417 ABM55423.1 AJWK01005764 GFTR01004066 JAW12360.1 GANO01001090 JAB58781.1 GBGD01002681 JAC86208.1 GECL01003422 JAP02702.1 GFDF01004631 JAV09453.1 KQ414617 KOC68382.1 ATLV01021599 KE525320 KFB46736.1 GBBI01000019 JAC18693.1 GAMD01002763 JAA98827.1 GGFM01003753 MBW24504.1 GGFK01006078 MBW39399.1 GGFJ01008770 MBW57911.1 GDKW01002194 JAI54401.1 ADMH02002149 ETN58251.1 AAAB01008978 EAA13600.2 APCN01002733 GDAI01001571 JAI16032.1 AXCM01006160 GG666475 EEN66630.1 GDIP01054323 GDIQ01030167 JAM49392.1 JAN64570.1 LRGB01002571 KZS07071.1 LJIJ01000018 ODN05707.1 NNAY01001464 OXU23883.1 AXCN02000110 CVRI01000059 CRL03349.1 GDIQ01039340 JAN55397.1 GDIP01203496 JAJ19906.1 GDIP01224840 JAI98561.1 GDIQ01157834 JAK93891.1 GDIP01190602 JAJ32800.1 GDIP01217646 JAJ05756.1 GL732612 EFX71218.1 GDIQ01214522 JAK37203.1 GDIP01199300 JAJ24102.1 GDIQ01269843 JAJ81881.1 GDIP01215303 JAJ08099.1 KK856463 PTY25826.1 GDIP01227295 JAI96106.1 GDIQ01178427 JAK73298.1 JW874556 AFP07073.1 GDIP01079412 JAM24303.1 JH432211 AMQN01001763 KB305378 ELU01128.1 GEDV01003722 JAP84835.1 JX209988 AFM88302.1
PCG76925.1 PCG76932.1 ODYU01006893 SOQ49147.1 GDQN01003648 JAT87406.1 AGBW02007696 OWR54697.1 RSAL01000034 RVE51372.1 GAIX01012328 JAA80232.1 KQ461185 KPJ07421.1 AK402517 BAM19139.1 AK401945 KQ459252 BAM18567.1 KPJ02194.1 NEVH01009101 PNF33583.1 KQ978411 KYM94100.1 GL452770 EFN76820.1 KQ978858 KYN27927.1 GL888206 EGI65134.1 ADTU01017554 KQ976453 KYM85393.1 KQ983247 KYQ46160.1 GECU01011980 JAS95726.1 GECZ01000676 JAS69093.1 GL439967 EFN66507.1 KK107078 QOIP01000011 EZA60401.1 RLU16699.1 LBMM01011888 KMQ86570.1 GEDC01001697 JAS35601.1 KQ981703 KYN37439.1 KC571887 AGM32386.1 GL767674 EFZ13292.1 KZ288363 PBC26923.1 GFDL01008793 JAV26252.1 DS231831 EDS31306.1 KK852869 KDR14657.1 CH477622 EAT38162.1 DS235283 EEB14389.1 JXUM01049009 KQ561586 KXJ78094.1 GAPW01003710 JAC09888.1 ACPB03006555 GAHY01000637 JAA76873.1 HP429363 ADN29863.1 EF179417 ABM55423.1 AJWK01005764 GFTR01004066 JAW12360.1 GANO01001090 JAB58781.1 GBGD01002681 JAC86208.1 GECL01003422 JAP02702.1 GFDF01004631 JAV09453.1 KQ414617 KOC68382.1 ATLV01021599 KE525320 KFB46736.1 GBBI01000019 JAC18693.1 GAMD01002763 JAA98827.1 GGFM01003753 MBW24504.1 GGFK01006078 MBW39399.1 GGFJ01008770 MBW57911.1 GDKW01002194 JAI54401.1 ADMH02002149 ETN58251.1 AAAB01008978 EAA13600.2 APCN01002733 GDAI01001571 JAI16032.1 AXCM01006160 GG666475 EEN66630.1 GDIP01054323 GDIQ01030167 JAM49392.1 JAN64570.1 LRGB01002571 KZS07071.1 LJIJ01000018 ODN05707.1 NNAY01001464 OXU23883.1 AXCN02000110 CVRI01000059 CRL03349.1 GDIQ01039340 JAN55397.1 GDIP01203496 JAJ19906.1 GDIP01224840 JAI98561.1 GDIQ01157834 JAK93891.1 GDIP01190602 JAJ32800.1 GDIP01217646 JAJ05756.1 GL732612 EFX71218.1 GDIQ01214522 JAK37203.1 GDIP01199300 JAJ24102.1 GDIQ01269843 JAJ81881.1 GDIP01215303 JAJ08099.1 KK856463 PTY25826.1 GDIP01227295 JAI96106.1 GDIQ01178427 JAK73298.1 JW874556 AFP07073.1 GDIP01079412 JAM24303.1 JH432211 AMQN01001763 KB305378 ELU01128.1 GEDV01003722 JAP84835.1 JX209988 AFM88302.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000235965 UP000078542 UP000008237 UP000078492 UP000007755 UP000005205 UP000078540 UP000075809 UP000000311 UP000002358 UP000053097 UP000279307 UP000036403 UP000078541 UP000242457 UP000002320 UP000027135 UP000008820 UP000009046 UP000069940 UP000249989 UP000015103 UP000092461 UP000075885 UP000053825 UP000030765 UP000069272 UP000075880 UP000075903 UP000000673 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075884 UP000076408 UP000075883 UP000075881 UP000001554 UP000085678 UP000076858 UP000094527 UP000215335 UP000075886 UP000075900 UP000183832 UP000075920 UP000000305 UP000014760
UP000235965 UP000078542 UP000008237 UP000078492 UP000007755 UP000005205 UP000078540 UP000075809 UP000000311 UP000002358 UP000053097 UP000279307 UP000036403 UP000078541 UP000242457 UP000002320 UP000027135 UP000008820 UP000009046 UP000069940 UP000249989 UP000015103 UP000092461 UP000075885 UP000053825 UP000030765 UP000069272 UP000075880 UP000075903 UP000000673 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075884 UP000076408 UP000075883 UP000075881 UP000001554 UP000085678 UP000076858 UP000094527 UP000215335 UP000075886 UP000075900 UP000183832 UP000075920 UP000000305 UP000014760
Interpro
SUPFAM
SSF56235
SSF56235
Gene 3D
ProteinModelPortal
Q1HPR1
H9J567
A0A2W1BTA5
A0A2A4JY31
A0A2H1W7W9
A0A1E1WKL4
+ More
A0A212FLT9 A0A3S2PH76 S4PTB5 A0A194QV38 I4DMJ9 I4DKX7 A0A2J7QYC4 A0A151I7N5 E2C5K7 A0A151JNM4 F4WKP2 A0A158NJ80 A0A195BJP3 A0A151WEA7 A0A1B6J933 A0A1B6H343 E2AJ67 K7ISL4 A0A026WXD0 A0A0J7N1R1 A0A1B6ECE7 A0A151JV70 R4UVY8 E9J1R8 A0A2A3E585 A0A1Q3FFG7 B0W3G3 A0A067R5N5 Q16UH5 E0VLY3 A0A182G942 A0A023EL07 R4G8P0 E2J7E5 A2IAA3 A0A1B0CBQ7 A0A224XVB3 U5EX64 A0A069DQ64 A0A0V0G3S8 A0A182PK35 A0A1L8DSK5 A0A0L7RBS3 A0A084W942 A0A023FCW9 T1E8H5 A0A2M3Z7L5 A0A2M4AF32 A0A2M4BYA8 A0A182FNY3 A0A0P4VNA4 A0A182J2G1 A0A182VAX2 W5J336 A0A182U7E3 A0A182XH14 A0A182KMV9 Q7Q2B0 A0A182I8D9 A0A182N6S9 A0A182XXL2 A0A0K8TNR5 A0A182M649 A0A182KFJ6 C3XZ66 A0A0P5YRK1 A0A1S3K3R9 A0A164PR99 A0A1D2NKE6 A0A232F000 A0A182Q3Z1 A0A182RY65 A0A1J1ISY9 A0A182VQG5 A0A0N8ECU1 A0A0P5AIU9 A0A0P4YNB4 A0A0P5MC31 A0A0P5BCN4 A0A0P4ZF56 E9HAP5 A0A0P5IEI8 A0A0P5ASX6 A0A0N8AJF2 A0A0P4ZB74 A0A2R7X246 A0A0P4YTZ2 A0A0N8BH29 V9L6Y5 A0A0P5WZX1 T1JGS3 R7UBE7 A0A131Z275 K4GK00
A0A212FLT9 A0A3S2PH76 S4PTB5 A0A194QV38 I4DMJ9 I4DKX7 A0A2J7QYC4 A0A151I7N5 E2C5K7 A0A151JNM4 F4WKP2 A0A158NJ80 A0A195BJP3 A0A151WEA7 A0A1B6J933 A0A1B6H343 E2AJ67 K7ISL4 A0A026WXD0 A0A0J7N1R1 A0A1B6ECE7 A0A151JV70 R4UVY8 E9J1R8 A0A2A3E585 A0A1Q3FFG7 B0W3G3 A0A067R5N5 Q16UH5 E0VLY3 A0A182G942 A0A023EL07 R4G8P0 E2J7E5 A2IAA3 A0A1B0CBQ7 A0A224XVB3 U5EX64 A0A069DQ64 A0A0V0G3S8 A0A182PK35 A0A1L8DSK5 A0A0L7RBS3 A0A084W942 A0A023FCW9 T1E8H5 A0A2M3Z7L5 A0A2M4AF32 A0A2M4BYA8 A0A182FNY3 A0A0P4VNA4 A0A182J2G1 A0A182VAX2 W5J336 A0A182U7E3 A0A182XH14 A0A182KMV9 Q7Q2B0 A0A182I8D9 A0A182N6S9 A0A182XXL2 A0A0K8TNR5 A0A182M649 A0A182KFJ6 C3XZ66 A0A0P5YRK1 A0A1S3K3R9 A0A164PR99 A0A1D2NKE6 A0A232F000 A0A182Q3Z1 A0A182RY65 A0A1J1ISY9 A0A182VQG5 A0A0N8ECU1 A0A0P5AIU9 A0A0P4YNB4 A0A0P5MC31 A0A0P5BCN4 A0A0P4ZF56 E9HAP5 A0A0P5IEI8 A0A0P5ASX6 A0A0N8AJF2 A0A0P4ZB74 A0A2R7X246 A0A0P4YTZ2 A0A0N8BH29 V9L6Y5 A0A0P5WZX1 T1JGS3 R7UBE7 A0A131Z275 K4GK00
PDB
6EPF
E-value=3.4886e-104,
Score=964
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
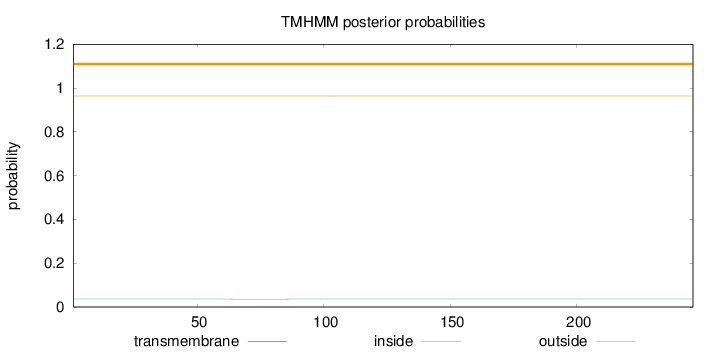
Length:
246
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01992
Exp number, first 60 AAs:
0.00148
Total prob of N-in:
0.03609
outside
1 - 246
Population Genetic Test Statistics
Pi
16.2283
Theta
19.112465
Tajima's D
-0.774893
CLR
1.174943
CSRT
0.186690665466727
Interpretation
Uncertain
