Pre Gene Modal
BGIBMGA004666
Annotation
small_GTP-binding_protein_Rab10_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.924
Sequence
CDS
ATGGCCAAAAAAACATACGACTTGTTATTCAAACTGCTTCTCATCGGTGACTCCGGAGTTGGAAAAACGTGCATATTGTTTAGGTTCTCAGATAACCACTTCACAACGACTTTTATTTCGACGATAGGTATCGACTTCAAAATAAAGACAGTGGAATTAAGGGGTAAGAAAATAAAGCTCCAGATATGGGACACTGCTGGGCAGGAGCGTTTCCACACCATCACGACGTCGTATTACCGCGGCGCTATGGGCATCATGCTCGTATACGACATAACAAATGAAAAAACATTTGATGATATCGTTAAATGGTTACGAAATATAGACGAGCATGCAAACGAGGATGTGGAAAAGATGATACTTGGCAATAAATGTGATATGGAGGAGAAACGTGTTGTCAGCAAGGAGCGCGGTGAAGCAATAGCCCGCGAGCACGGCATCCGCTTCATGGAGACGTCGGCTAAAGCGAACATAAATATAGACAGAGCTTTCTCGGAACTAGCGGAGGCTATACTCGACAAGACGGCGGGGCGCGAGGCAGACGCCGACCACTCCCGCATCGCGGTCGAGCGCCGCCCCTCCCGCGCCCCGCCCTCGCGCGCCTGCTGCACATAG
Protein
MAKKTYDLLFKLLLIGDSGVGKTCILFRFSDNHFTTTFISTIGIDFKIKTVELRGKKIKLQIWDTAGQERFHTITTSYYRGAMGIMLVYDITNEKTFDDIVKWLRNIDEHANEDVEKMILGNKCDMEEKRVVSKERGEAIAREHGIRFMETSAKANINIDRAFSELAEAILDKTAGREADADHSRIAVERRPSRAPPSRACCT
Summary
Uniprot
Q0N2S2
A0A2A4JZU3
A0A2H1W7T4
A0A194QPG3
A0A2W1BZL4
A0A212FLX1
+ More
A0A194Q9J9 A0A1A9Y8M3 A0A1A9UHY9 A0A1B0C3V9 A0A1A9ZBC2 D3TNT5 A0A1A9W935 A0A0L0C293 A0A1W4XUG1 A0A026WQJ3 F4W459 A0A195B3F1 A0A195ETC1 A0A158NF30 A0A151X1W9 A0A1Y1N6E8 B4NCN8 A0A154PCA5 B4M9Y7 A0A195CLK5 E2AT08 B4L7W8 B4PY14 B4I654 B3NWH1 O15971 E9I9V7 A0A067R747 A0A3B0K0F3 A0A0C9RTU9 A0A2J7PJS8 D6WHE1 A0A310STY6 A0A0N0U6J3 A0A087ZVR2 A0A1I8QCX8 B4JKA5 A0A1I8NAV0 A0A2A3EAQ6 A0A0M5J8A4 A0A034WV28 W8AGQ9 B5DN66 A0A1W4VG45 E2B6P6 A0A0T6AXI3 A0A0K8UJI2 A0A0K8TNH4 A0A1B6M051 A0A1B6F8F5 A0A023EJH6 A0A1Q3FWN5 A0A0L7QSX5 U5EZM3 Q17J36 A0A0J7L967 B3MQI0 A0A182FIL5 A0A1L8EH58 A0A1J1J4L1 A0A1B0CFI1 A0A1L8DVH3 K7INY9 A0A232F6K3 A0A336MMZ4 A0A0B4UBW3 A0A182NAD2 A0A084WPI3 A0A182JBA7 A0A2M4C024 A0A182QHP3 N6U250 A0A2M3ZJ48 W5JMM4 A0A182X7G4 A0A182V6E8 A0A182U510 A0A182LDM8 Q7QEG1 A0A182I9X5 A0A0A9WQ73 A0A2M4AQS5 T1D4Q7 A0A182M537 A0A1J1HNH7 A0A0V0G7B7 A0A023FAU6 A0A069DPZ5 A0A0P4VUC0 R4G8M2 A0A3R7M456 A0A0P4W546 T1DF43 E9HIB2
A0A194Q9J9 A0A1A9Y8M3 A0A1A9UHY9 A0A1B0C3V9 A0A1A9ZBC2 D3TNT5 A0A1A9W935 A0A0L0C293 A0A1W4XUG1 A0A026WQJ3 F4W459 A0A195B3F1 A0A195ETC1 A0A158NF30 A0A151X1W9 A0A1Y1N6E8 B4NCN8 A0A154PCA5 B4M9Y7 A0A195CLK5 E2AT08 B4L7W8 B4PY14 B4I654 B3NWH1 O15971 E9I9V7 A0A067R747 A0A3B0K0F3 A0A0C9RTU9 A0A2J7PJS8 D6WHE1 A0A310STY6 A0A0N0U6J3 A0A087ZVR2 A0A1I8QCX8 B4JKA5 A0A1I8NAV0 A0A2A3EAQ6 A0A0M5J8A4 A0A034WV28 W8AGQ9 B5DN66 A0A1W4VG45 E2B6P6 A0A0T6AXI3 A0A0K8UJI2 A0A0K8TNH4 A0A1B6M051 A0A1B6F8F5 A0A023EJH6 A0A1Q3FWN5 A0A0L7QSX5 U5EZM3 Q17J36 A0A0J7L967 B3MQI0 A0A182FIL5 A0A1L8EH58 A0A1J1J4L1 A0A1B0CFI1 A0A1L8DVH3 K7INY9 A0A232F6K3 A0A336MMZ4 A0A0B4UBW3 A0A182NAD2 A0A084WPI3 A0A182JBA7 A0A2M4C024 A0A182QHP3 N6U250 A0A2M3ZJ48 W5JMM4 A0A182X7G4 A0A182V6E8 A0A182U510 A0A182LDM8 Q7QEG1 A0A182I9X5 A0A0A9WQ73 A0A2M4AQS5 T1D4Q7 A0A182M537 A0A1J1HNH7 A0A0V0G7B7 A0A023FAU6 A0A069DPZ5 A0A0P4VUC0 R4G8M2 A0A3R7M456 A0A0P4W546 T1DF43 E9HIB2
Pubmed
19121390
26354079
28756777
22118469
20353571
26108605
+ More
24508170 30249741 21719571 21347285 28004739 17994087 20798317 17550304 9074639 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 24845553 18362917 19820115 25315136 25348373 24495485 15632085 26369729 24945155 26483478 17510324 20075255 28648823 25542378 24438588 23537049 20920257 23761445 20966253 12364791 14747013 17210077 25401762 26823975 24330624 25474469 26334808 27129103 21292972
24508170 30249741 21719571 21347285 28004739 17994087 20798317 17550304 9074639 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 24845553 18362917 19820115 25315136 25348373 24495485 15632085 26369729 24945155 26483478 17510324 20075255 28648823 25542378 24438588 23537049 20920257 23761445 20966253 12364791 14747013 17210077 25401762 26823975 24330624 25474469 26334808 27129103 21292972
EMBL
BABH01039391
DQ646409
ABH10799.1
NWSH01000369
PCG76930.1
ODYU01006893
+ More
SOQ49149.1 KQ461185 KPJ07423.1 KZ149906 PZC78300.1 AGBW02007696 OWR54699.1 KQ459252 KPJ02192.1 JXJN01025121 CCAG010013941 EZ423087 ADD19363.1 JRES01000984 KNC26473.1 KK107131 QOIP01000012 EZA58208.1 RLU16268.1 GL887491 EGI71055.1 KQ976625 KYM79016.1 KQ981979 KYN31483.1 ADTU01001500 KQ982585 KYQ54285.1 GEZM01011831 GEZM01011830 JAV93421.1 CH964239 EDW82597.1 KQ434870 KZC09525.1 CH940655 EDW66046.1 KQ977642 KYN00954.1 GL442439 EFN63443.1 CH933814 EDW05543.1 CM000162 EDX02986.1 CH480823 EDW56260.1 CH954180 EDV46791.1 AE014298 AY060425 AB006189 AAF50924.1 AAL25464.1 BAA21744.1 GL761846 EFZ22660.1 KK852833 KDR15303.1 OUUW01000003 SPP78491.1 GBYB01012160 JAG81927.1 NEVH01024946 PNF16559.1 KQ971321 EFA00103.1 KQ760208 OAD61379.1 KQ435727 KOX78078.1 CH916370 EDW00008.1 KZ288304 PBC28798.1 CP012528 ALC49381.1 GAKP01001324 JAC57628.1 GAMC01019002 GAMC01019000 JAB87555.1 CH379064 EDY72873.1 GL446000 EFN88630.1 LJIG01022608 KRT79659.1 GDHF01025462 JAI26852.1 GDAI01001661 JAI15942.1 GEBQ01010676 JAT29301.1 GECZ01027922 GECZ01023338 GECZ01020028 JAS41847.1 JAS46431.1 JAS49741.1 JXUM01099004 GAPW01004483 KQ564462 JAC09115.1 KXJ72236.1 GFDL01003087 JAV31958.1 KQ414755 KOC61742.1 GANO01000527 JAB59344.1 CH477235 EAT46674.1 EJY57406.1 LBMM01000221 KMR04267.1 CH902621 EDV44606.1 GFDG01000778 JAV18021.1 CVRI01000065 CRL05809.1 AJWK01010107 GFDF01003789 JAV10295.1 NNAY01000833 OXU26275.1 UFQT01001779 SSX31772.1 KP216760 AJC97117.1 ATLV01025079 KE525369 KFB52127.1 AXCP01007413 GGFJ01009463 MBW58604.1 AXCN02001450 APGK01042609 KB741007 KB632182 ENN75610.1 ERL89617.1 GGFM01007744 MBW28495.1 ADMH02001131 ETN64009.1 AAAB01008847 EAA06827.3 APCN01001073 GBHO01034991 GBRD01002698 GDHC01003916 JAG08613.1 JAG63123.1 JAQ14713.1 GGFK01009802 MBW43123.1 GALA01000853 JAA93999.1 AXCM01006226 CVRI01000013 CRK89607.1 GECL01002106 JAP04018.1 GBBI01000558 JAC18154.1 GBGD01003004 JAC85885.1 GDKW01000967 JAI55628.1 ACPB03004243 GAHY01000727 JAA76783.1 QCYY01002892 ROT66744.1 GDRN01091327 JAI60291.1 GALA01000797 JAA94055.1 GL732654 EFX68468.1
SOQ49149.1 KQ461185 KPJ07423.1 KZ149906 PZC78300.1 AGBW02007696 OWR54699.1 KQ459252 KPJ02192.1 JXJN01025121 CCAG010013941 EZ423087 ADD19363.1 JRES01000984 KNC26473.1 KK107131 QOIP01000012 EZA58208.1 RLU16268.1 GL887491 EGI71055.1 KQ976625 KYM79016.1 KQ981979 KYN31483.1 ADTU01001500 KQ982585 KYQ54285.1 GEZM01011831 GEZM01011830 JAV93421.1 CH964239 EDW82597.1 KQ434870 KZC09525.1 CH940655 EDW66046.1 KQ977642 KYN00954.1 GL442439 EFN63443.1 CH933814 EDW05543.1 CM000162 EDX02986.1 CH480823 EDW56260.1 CH954180 EDV46791.1 AE014298 AY060425 AB006189 AAF50924.1 AAL25464.1 BAA21744.1 GL761846 EFZ22660.1 KK852833 KDR15303.1 OUUW01000003 SPP78491.1 GBYB01012160 JAG81927.1 NEVH01024946 PNF16559.1 KQ971321 EFA00103.1 KQ760208 OAD61379.1 KQ435727 KOX78078.1 CH916370 EDW00008.1 KZ288304 PBC28798.1 CP012528 ALC49381.1 GAKP01001324 JAC57628.1 GAMC01019002 GAMC01019000 JAB87555.1 CH379064 EDY72873.1 GL446000 EFN88630.1 LJIG01022608 KRT79659.1 GDHF01025462 JAI26852.1 GDAI01001661 JAI15942.1 GEBQ01010676 JAT29301.1 GECZ01027922 GECZ01023338 GECZ01020028 JAS41847.1 JAS46431.1 JAS49741.1 JXUM01099004 GAPW01004483 KQ564462 JAC09115.1 KXJ72236.1 GFDL01003087 JAV31958.1 KQ414755 KOC61742.1 GANO01000527 JAB59344.1 CH477235 EAT46674.1 EJY57406.1 LBMM01000221 KMR04267.1 CH902621 EDV44606.1 GFDG01000778 JAV18021.1 CVRI01000065 CRL05809.1 AJWK01010107 GFDF01003789 JAV10295.1 NNAY01000833 OXU26275.1 UFQT01001779 SSX31772.1 KP216760 AJC97117.1 ATLV01025079 KE525369 KFB52127.1 AXCP01007413 GGFJ01009463 MBW58604.1 AXCN02001450 APGK01042609 KB741007 KB632182 ENN75610.1 ERL89617.1 GGFM01007744 MBW28495.1 ADMH02001131 ETN64009.1 AAAB01008847 EAA06827.3 APCN01001073 GBHO01034991 GBRD01002698 GDHC01003916 JAG08613.1 JAG63123.1 JAQ14713.1 GGFK01009802 MBW43123.1 GALA01000853 JAA93999.1 AXCM01006226 CVRI01000013 CRK89607.1 GECL01002106 JAP04018.1 GBBI01000558 JAC18154.1 GBGD01003004 JAC85885.1 GDKW01000967 JAI55628.1 ACPB03004243 GAHY01000727 JAA76783.1 QCYY01002892 ROT66744.1 GDRN01091327 JAI60291.1 GALA01000797 JAA94055.1 GL732654 EFX68468.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000092443
+ More
UP000078200 UP000092460 UP000092445 UP000092444 UP000091820 UP000037069 UP000192223 UP000053097 UP000279307 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000007798 UP000076502 UP000008792 UP000078542 UP000000311 UP000009192 UP000002282 UP000001292 UP000008711 UP000000803 UP000027135 UP000268350 UP000235965 UP000007266 UP000053105 UP000005203 UP000095300 UP000001070 UP000095301 UP000242457 UP000092553 UP000001819 UP000192221 UP000008237 UP000069940 UP000249989 UP000053825 UP000008820 UP000036403 UP000007801 UP000069272 UP000183832 UP000092461 UP000002358 UP000215335 UP000075884 UP000030765 UP000075880 UP000075886 UP000019118 UP000030742 UP000000673 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000075883 UP000015103 UP000283509 UP000000305
UP000078200 UP000092460 UP000092445 UP000092444 UP000091820 UP000037069 UP000192223 UP000053097 UP000279307 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000007798 UP000076502 UP000008792 UP000078542 UP000000311 UP000009192 UP000002282 UP000001292 UP000008711 UP000000803 UP000027135 UP000268350 UP000235965 UP000007266 UP000053105 UP000005203 UP000095300 UP000001070 UP000095301 UP000242457 UP000092553 UP000001819 UP000192221 UP000008237 UP000069940 UP000249989 UP000053825 UP000008820 UP000036403 UP000007801 UP000069272 UP000183832 UP000092461 UP000002358 UP000215335 UP000075884 UP000030765 UP000075880 UP000075886 UP000019118 UP000030742 UP000000673 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000075883 UP000015103 UP000283509 UP000000305
Interpro
Gene 3D
ProteinModelPortal
Q0N2S2
A0A2A4JZU3
A0A2H1W7T4
A0A194QPG3
A0A2W1BZL4
A0A212FLX1
+ More
A0A194Q9J9 A0A1A9Y8M3 A0A1A9UHY9 A0A1B0C3V9 A0A1A9ZBC2 D3TNT5 A0A1A9W935 A0A0L0C293 A0A1W4XUG1 A0A026WQJ3 F4W459 A0A195B3F1 A0A195ETC1 A0A158NF30 A0A151X1W9 A0A1Y1N6E8 B4NCN8 A0A154PCA5 B4M9Y7 A0A195CLK5 E2AT08 B4L7W8 B4PY14 B4I654 B3NWH1 O15971 E9I9V7 A0A067R747 A0A3B0K0F3 A0A0C9RTU9 A0A2J7PJS8 D6WHE1 A0A310STY6 A0A0N0U6J3 A0A087ZVR2 A0A1I8QCX8 B4JKA5 A0A1I8NAV0 A0A2A3EAQ6 A0A0M5J8A4 A0A034WV28 W8AGQ9 B5DN66 A0A1W4VG45 E2B6P6 A0A0T6AXI3 A0A0K8UJI2 A0A0K8TNH4 A0A1B6M051 A0A1B6F8F5 A0A023EJH6 A0A1Q3FWN5 A0A0L7QSX5 U5EZM3 Q17J36 A0A0J7L967 B3MQI0 A0A182FIL5 A0A1L8EH58 A0A1J1J4L1 A0A1B0CFI1 A0A1L8DVH3 K7INY9 A0A232F6K3 A0A336MMZ4 A0A0B4UBW3 A0A182NAD2 A0A084WPI3 A0A182JBA7 A0A2M4C024 A0A182QHP3 N6U250 A0A2M3ZJ48 W5JMM4 A0A182X7G4 A0A182V6E8 A0A182U510 A0A182LDM8 Q7QEG1 A0A182I9X5 A0A0A9WQ73 A0A2M4AQS5 T1D4Q7 A0A182M537 A0A1J1HNH7 A0A0V0G7B7 A0A023FAU6 A0A069DPZ5 A0A0P4VUC0 R4G8M2 A0A3R7M456 A0A0P4W546 T1DF43 E9HIB2
A0A194Q9J9 A0A1A9Y8M3 A0A1A9UHY9 A0A1B0C3V9 A0A1A9ZBC2 D3TNT5 A0A1A9W935 A0A0L0C293 A0A1W4XUG1 A0A026WQJ3 F4W459 A0A195B3F1 A0A195ETC1 A0A158NF30 A0A151X1W9 A0A1Y1N6E8 B4NCN8 A0A154PCA5 B4M9Y7 A0A195CLK5 E2AT08 B4L7W8 B4PY14 B4I654 B3NWH1 O15971 E9I9V7 A0A067R747 A0A3B0K0F3 A0A0C9RTU9 A0A2J7PJS8 D6WHE1 A0A310STY6 A0A0N0U6J3 A0A087ZVR2 A0A1I8QCX8 B4JKA5 A0A1I8NAV0 A0A2A3EAQ6 A0A0M5J8A4 A0A034WV28 W8AGQ9 B5DN66 A0A1W4VG45 E2B6P6 A0A0T6AXI3 A0A0K8UJI2 A0A0K8TNH4 A0A1B6M051 A0A1B6F8F5 A0A023EJH6 A0A1Q3FWN5 A0A0L7QSX5 U5EZM3 Q17J36 A0A0J7L967 B3MQI0 A0A182FIL5 A0A1L8EH58 A0A1J1J4L1 A0A1B0CFI1 A0A1L8DVH3 K7INY9 A0A232F6K3 A0A336MMZ4 A0A0B4UBW3 A0A182NAD2 A0A084WPI3 A0A182JBA7 A0A2M4C024 A0A182QHP3 N6U250 A0A2M3ZJ48 W5JMM4 A0A182X7G4 A0A182V6E8 A0A182U510 A0A182LDM8 Q7QEG1 A0A182I9X5 A0A0A9WQ73 A0A2M4AQS5 T1D4Q7 A0A182M537 A0A1J1HNH7 A0A0V0G7B7 A0A023FAU6 A0A069DPZ5 A0A0P4VUC0 R4G8M2 A0A3R7M456 A0A0P4W546 T1DF43 E9HIB2
PDB
5SZJ
E-value=3.23323e-82,
Score=773
Ontologies
GO
GO:0005525
GO:0003924
GO:0007295
GO:0110010
GO:0045175
GO:0110011
GO:0045178
GO:0016192
GO:0005737
GO:0031254
GO:0061864
GO:0016328
GO:0032482
GO:0045202
GO:0031982
GO:0055037
GO:0043025
GO:0032869
GO:0005768
GO:0072659
GO:0006886
GO:0017157
GO:0009306
GO:0005794
GO:0005886
GO:0006904
GO:0008021
GO:0032593
GO:0007264
GO:0005515
GO:0004888
GO:0043565
GO:0043401
GO:0006418
GO:0016020
GO:0005634
GO:0006260
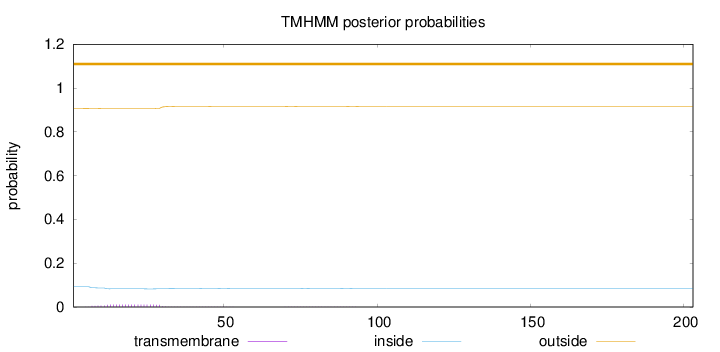
Topology
Length:
203
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26234
Exp number, first 60 AAs:
0.24243
Total prob of N-in:
0.09249
outside
1 - 203
Population Genetic Test Statistics
Pi
5.723842
Theta
15.216663
Tajima's D
-1.09805
CLR
17.731144
CSRT
0.115694215289236
Interpretation
Uncertain
