Pre Gene Modal
BGIBMGA004658
Annotation
putative_stromal_antigen_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 2.64
Sequence
CDS
ATGCATCGTCGAGGCGGGAAACGTATACGGATGGACGATCCTCCACCGGAGTATGTAAATCCTATGACTCCGGCTACTCCTATGACGGACTATGGTGGCCAATCTGTCCATGAACCTGAAACGCCTAATGTGAACTATAGTGGCTTTAATACTGGTGCAGTAGCTAACTCGACGGCAGTGGAAGCGCCACGAGAGGAAGCTGAAGAATCGCATCACGAGGAAGAGGAACCGAGGTCTCCATCTCCGACCCTCAAGCGGGTGACTCGAAGTCGAGGTAGACGCGCAGATGGTGGTTATGTGGGGCGATTGGCGGAGTCTCCGCCGCCACCGCCTATACGACGTCGAGGCCGCGGTGTCCGTGGTCGTGGCAGAGGACGAGGTGCCGCTCCACCTCCAGCATATTCTCCACCCCCGGTGCTCCTGCCTGGAGATGATGAAAACAGTTTATATAATATTTTGAGGTTCAACAAGACCGCAATAAACCAAGTAGTAGACAAATGGATAGAGGAGTACAAAACAAATCGTGAAGTGGCTTTAGTTCAACTGATGCAGTTCTTCATCAACTCTTCCGGTTGCAGAGGGAAGGTTTCTCCAGATATGGCACAGATGGATCACACACTAATAATAAAAAAAATGACACAGGAATTTGATGAGGAAAGTGGTGAATATCCTCTGATAATGACTGGTCAAACCTGGAAAAAGTTCCGTTCTAACTTTTGTGAGTTCATTCAGACCCTGGTAAAGATGTGCCAATACTCCATCATCTATGATCAATATTTAATGGACAATATCATTTCATTACTCACTGGGCTGTCTGACTCACAAGTCCGAGCTTTCCGTCACACTGCGACATTAGCTGTAATGAAGCTAATGACCGCTCTAGTTGATGTGGCGTTATTAACTAGTGTAAATTGTGACAACTGTCTTCGGCAATATGAAGCAGAAAGATTGAAGGCACGTGATAAGCGGGCCACTGAACGCCTTGAAGTGCTGGTGGCTAAACGTCAAGAACTAGAGGAGAACATGGAGGAGATTAAGAATATGCTGTCTTATATGTTTAAATCTGTGTTTGTCCATCGATACAGGGACACTTTGCCAGATATAAGAGCCATAACTATGTCTGAGATTGGCATTTGGATGGAGAAATTTCCAGCTCATTTCCTTGATGATCTTTATTTAAAATATATTGGATGGACACTTCATGATAAGGTGGGTGAAGTGCGTCTTCGCTGTCTGCAAGCCCTACAACCTCTATATGAATGTGAGGAGTTGAAGGGTAAACTGGAACTCTTCACGTCTAAATTCAAAGATCGGATAGTTTCTATGACCCTCGATAAGGAAACTGATGTGGCCGTGCATGCTGTGAAGTTGGTTATAGCTATTCTCAAGATGCACCCGGACGTGCTGACGGACAAGGACTGCGAGAACGTGTACGAACTGGTGTACTCGTCGTGGCGCGGCGTGGCGGCGGCGGCCGGAGAGTTCCTCAACGTGCGCCTGTTCCGCGCCGACCCCGCCGCGCCCGCGCCCCCCGTGCTGCGCTCGCGCCGCGGCAAGCTGCGCCTGCCCAGCACGCCGCTCGTGCGGGACCTCGTGCAGTTCTTCATAGAGTCCGAGTTGCATGAACATGGTGCTTATTTAGTGGATTCATTGATTGAATCAAATCCCATGATGAAGGATTGGGAGTGTATGACTGACCTACTGCTCGAAGAAGCTGGTCCTGGGGAAGAGGCACTGGATAATAGACAGGAGTCGTCGCTGATTGAGCTGCTGGTGTGCTGCGTGCGGCAGGCGTCCACCGGGGAGCCGCCCGTGGGCCGCCAGCCCTCGCGCAAGCAGCACGTCGCGCTCTCCAAGGATCAGGCCAAGGTTGTAAGCGACGACCGAGCTAAAATGACAGCACACTTCATTGTGACCCTGCCGGCCCTGCTGGACAAGTTCGGTGCGGACCCGGAGAAGCTCACCAACCTGGTGTCCATCCCGCAGTACTTCGACCTGGAGATCTACACGACGCAACGGCAGGAAGGGAACCTAACCCTGCTGCTGAACAAGCTGCGCGACATCGTGAGCGTGCAGACGGAGGCGGAGGTGCTGGAGACGTGCGGGCGCACGCTGGAGTTCCTGTGCAGCGAGGGCTGCTCCGTGTACACGCGCTGCAACGTCGCGCGCGCCACCATCACCGACGCCTGCGTCAACCGATACAAGGAGGCCATCGACGACTACAGGAGCCTCATCGAAGGGGGTGAAACACCGGATGCAGATGAAGTGTTCAACGTGATAAATTCCCTGCGGAAAGTTTCCATAATGTACATGTGTCACAATCTGAACGACACCAATATCTGGGACTCGCTGTTTGAAGACCTACCCAAGTGCTTGAAGCAGAATGAAACACAGATGCCGTCACAGGCCCTGGTGTACGTGGTGCGCGCGCTGTTCTACTCCGTGCTGTGGTCGCTGCACGCGCTGGAGGAGCGCGCGGGGGGTGCGGGCGGCGCGGGGGGTGGCGCGGGGGGAGCGGGGGGCGCGGCGGCGCTGCGGGACCGGCTGCGCGCCTACTGCGCTCTGTGCACGGACATCGTCGTCAACGGGACCACGCCGGAGCTCAAGGAGGAGGCCTACACGAGCATCTGCGACCTGCTGATATTCTTCGCGGAGCACCTGCACACGCTGCACCACCCCAACGAGCCGTCCATGAAGCAGCTGGTGTACCAGCCCGACAACGACCTGTGCGACTTGCTCAACGACTTCATACAGGAATTCGTGTTTGTACAACACAATTACGATGGTCAAGATGAGAGACGCATAGAGGAACTGCACAAGCGCCGCAACTTCCTGGCGGCCTACTGTAAGCTCATCGTGTACAACGTGGCGCCCATACGCCGCGCCGCGGACGTCTTCAAACACTACATCAAGTGCTACAACGACTACGGCGACATAATCAAGGCGACGCTGAGCAAGGCGCGCGAGATCAACAAGCTGAACTGCGCGCTCACCATGGAGCTGGCCATGCAGGCGCTGTTCCGCGACCTGCTGCGCCGCCACGCCACCCCGCACCGACAGCTGCCGGAGTTCCTCGAGCTCAAGGAGCTGGCGAAGCGTTTCTCCGTGATGTTCGGTCTGGACGCGGTGAAGAACCGCGAGGCGCTGACCGCGCTGCACCGCGCCGGCATCGCCTTCGCCGCGCTCGACCCCGCGCCGCACGCGCCGCCGCCCAACCTGCTGTTCCTCGAGCCGCTCGGGGAGTTCTCCAACAAGCTGCTGCGCCAGGACAAGCGACAGGTGCTCAAGTTCCTGGAGTCTCGCATCCCGCAAGGGCTGCAGTGGGGCGAGGAGTGGACGGCGCTGGCGGCCTACCGCAGCGCGCTGCTCACCGACGCGCCGGACGAGCGCCCGCCGCCCGTGCGCAGGCACTACTCGCGGAGGGGACGACACGGTCAAAACGAAGACGAAGAAGGCGATGAGAACGCAGAATCGGACGGTGACCATCCAGGCACACATAAATGA
Protein
MHRRGGKRIRMDDPPPEYVNPMTPATPMTDYGGQSVHEPETPNVNYSGFNTGAVANSTAVEAPREEAEESHHEEEEPRSPSPTLKRVTRSRGRRADGGYVGRLAESPPPPPIRRRGRGVRGRGRGRGAAPPPAYSPPPVLLPGDDENSLYNILRFNKTAINQVVDKWIEEYKTNREVALVQLMQFFINSSGCRGKVSPDMAQMDHTLIIKKMTQEFDEESGEYPLIMTGQTWKKFRSNFCEFIQTLVKMCQYSIIYDQYLMDNIISLLTGLSDSQVRAFRHTATLAVMKLMTALVDVALLTSVNCDNCLRQYEAERLKARDKRATERLEVLVAKRQELEENMEEIKNMLSYMFKSVFVHRYRDTLPDIRAITMSEIGIWMEKFPAHFLDDLYLKYIGWTLHDKVGEVRLRCLQALQPLYECEELKGKLELFTSKFKDRIVSMTLDKETDVAVHAVKLVIAILKMHPDVLTDKDCENVYELVYSSWRGVAAAAGEFLNVRLFRADPAAPAPPVLRSRRGKLRLPSTPLVRDLVQFFIESELHEHGAYLVDSLIESNPMMKDWECMTDLLLEEAGPGEEALDNRQESSLIELLVCCVRQASTGEPPVGRQPSRKQHVALSKDQAKVVSDDRAKMTAHFIVTLPALLDKFGADPEKLTNLVSIPQYFDLEIYTTQRQEGNLTLLLNKLRDIVSVQTEAEVLETCGRTLEFLCSEGCSVYTRCNVARATITDACVNRYKEAIDDYRSLIEGGETPDADEVFNVINSLRKVSIMYMCHNLNDTNIWDSLFEDLPKCLKQNETQMPSQALVYVVRALFYSVLWSLHALEERAGGAGGAGGGAGGAGGAAALRDRLRAYCALCTDIVVNGTTPELKEEAYTSICDLLIFFAEHLHTLHHPNEPSMKQLVYQPDNDLCDLLNDFIQEFVFVQHNYDGQDERRIEELHKRRNFLAAYCKLIVYNVAPIRRAADVFKHYIKCYNDYGDIIKATLSKAREINKLNCALTMELAMQALFRDLLRRHATPHRQLPEFLELKELAKRFSVMFGLDAVKNREALTALHRAGIAFAALDPAPHAPPPNLLFLEPLGEFSNKLLRQDKRQVLKFLESRIPQGLQWGEEWTALAAYRSALLTDAPDERPPPVRRHYSRRGRHGQNEDEEGDENAESDGDHPGTHK
Summary
Uniprot
H9J568
A0A2H1W7V1
A0A212FLX5
A0A194QPY8
A0A2W1BTB9
A0A2A4JYP2
+ More
A0A194QFE5 A0A0T6B3F1 A0A2J7PZ83 A0A067QV78 D6WLF4 A0A1Y1LJL7 Q17D80 A0A0P6IVK3 A0A1S4F7G9 A0A291S6X8 A0A182G7H5 B0WRV8 A0A1Q3FCK6 A0A1Q3FCF5 N6UG33 A0A1B6E0R9 A0A151IH37 A0A1B6GWN1 A0A195E7H7 A0A1B6KVR2 A0A195F2E3 A0A158NLC3 F4X6M7 A0A195B1U8 A0A151XCB7 A0A1B6KZR0 A0A0J7LAK2 W8BQ30 A0A182FGC3 A0A2M4CYG0 A0A2M4CYU4 A0A182JTG5 A0A0A1XDL2 A0A182QQX9 A0A146LJ60 E2BMN9 A0A0L7QNJ8 A0A3L8DQ63 A0A034W1S0 A0A2A3ED52 A0A026WYN6 A0A182NJT9 A0A182LTU8 A0A1A9VIS1 A0A1B0A6D5 A0A1B0GAR0 A0A1A9X0J4 A0A182Y313 A0A182P135 A0A0L0BR21 A0A084WRQ4 A0A182L3H4 A0A182V4C5 A0A154PPB6 A0A182TRJ4 Q7PTA6 A0A182WWM4 A0A182I6W0 A0A1I8N2I6 A0A182JA52 A0A182WHX9 W5J562 T1PK51 A0A1I8N2I8 A0A1I8PJA4 A0A1I8PJD0 T1HNF5 A0A1I8PJ88 A0A1B0C0C7 A0A1I8PJ20 E0VQR7 A0A1W4UQH3 A0A0K8VLP5 B3MKS3 A0A088A791 A0A182R9V0 B4N064 B3N669 A0A0P6E9G5 A0A0N8ENM6 A0A1A9YKR0 A0A1B0C5B4 A0A0P5SFL2 E9FV02 A0A0P5QH58 A0A164PZ63 A0A0P6F3M4 B4NZY6 A0A0P4XU45 A0A0P5A1V2 A0A0P4Y1T3 A0A0P5R2A2 B4Q5A2 A0A0P6FHV1
A0A194QFE5 A0A0T6B3F1 A0A2J7PZ83 A0A067QV78 D6WLF4 A0A1Y1LJL7 Q17D80 A0A0P6IVK3 A0A1S4F7G9 A0A291S6X8 A0A182G7H5 B0WRV8 A0A1Q3FCK6 A0A1Q3FCF5 N6UG33 A0A1B6E0R9 A0A151IH37 A0A1B6GWN1 A0A195E7H7 A0A1B6KVR2 A0A195F2E3 A0A158NLC3 F4X6M7 A0A195B1U8 A0A151XCB7 A0A1B6KZR0 A0A0J7LAK2 W8BQ30 A0A182FGC3 A0A2M4CYG0 A0A2M4CYU4 A0A182JTG5 A0A0A1XDL2 A0A182QQX9 A0A146LJ60 E2BMN9 A0A0L7QNJ8 A0A3L8DQ63 A0A034W1S0 A0A2A3ED52 A0A026WYN6 A0A182NJT9 A0A182LTU8 A0A1A9VIS1 A0A1B0A6D5 A0A1B0GAR0 A0A1A9X0J4 A0A182Y313 A0A182P135 A0A0L0BR21 A0A084WRQ4 A0A182L3H4 A0A182V4C5 A0A154PPB6 A0A182TRJ4 Q7PTA6 A0A182WWM4 A0A182I6W0 A0A1I8N2I6 A0A182JA52 A0A182WHX9 W5J562 T1PK51 A0A1I8N2I8 A0A1I8PJA4 A0A1I8PJD0 T1HNF5 A0A1I8PJ88 A0A1B0C0C7 A0A1I8PJ20 E0VQR7 A0A1W4UQH3 A0A0K8VLP5 B3MKS3 A0A088A791 A0A182R9V0 B4N064 B3N669 A0A0P6E9G5 A0A0N8ENM6 A0A1A9YKR0 A0A1B0C5B4 A0A0P5SFL2 E9FV02 A0A0P5QH58 A0A164PZ63 A0A0P6F3M4 B4NZY6 A0A0P4XU45 A0A0P5A1V2 A0A0P4Y1T3 A0A0P5R2A2 B4Q5A2 A0A0P6FHV1
Pubmed
19121390
22118469
26354079
28756777
24845553
18362917
+ More
19820115 28004739 17510324 26999592 28992199 26483478 23537049 21347285 21719571 24495485 25830018 26823975 20798317 30249741 25348373 24508170 25244985 26108605 24438588 20966253 12364791 25315136 20920257 23761445 20566863 17994087 21292972 17550304 22936249
19820115 28004739 17510324 26999592 28992199 26483478 23537049 21347285 21719571 24495485 25830018 26823975 20798317 30249741 25348373 24508170 25244985 26108605 24438588 20966253 12364791 25315136 20920257 23761445 20566863 17994087 21292972 17550304 22936249
EMBL
BABH01039384
BABH01039385
BABH01039386
BABH01039387
BABH01039388
BABH01039389
+ More
BABH01039390 BABH01039391 ODYU01006893 SOQ49150.1 AGBW02007696 OWR54700.1 KQ461185 KPJ07424.1 KZ149906 PZC78299.1 NWSH01000369 PCG76929.1 KQ459252 KPJ02191.1 LJIG01015976 KRT81987.1 NEVH01020337 PNF21651.1 KK852909 KDR13941.1 KQ971343 EFA04673.2 GEZM01057701 JAV72135.1 CH477298 EAT44349.1 GDUN01000889 JAN95030.1 MF433013 ATL75364.1 JXUM01150325 KQ570973 KXJ68272.1 DS232061 EDS33523.1 GFDL01009731 JAV25314.1 GFDL01009774 JAV25271.1 APGK01028388 APGK01028389 KB740686 KB631879 ENN79576.1 ERL86904.1 GEDC01005773 JAS31525.1 KQ977647 KYN00840.1 GECZ01002928 JAS66841.1 KQ979568 KYN20789.1 GEBQ01024444 JAT15533.1 KQ981864 KYN34347.1 ADTU01019317 GL888818 EGI57809.1 KQ976662 KYM78448.1 KQ982314 KYQ58004.1 GEBQ01023040 JAT16937.1 LBMM01000035 KMR05231.1 GAMC01003200 JAC03356.1 GGFL01006194 MBW70372.1 GGFL01006193 MBW70371.1 GBXI01004808 JAD09484.1 AXCN02000522 GDHC01010456 JAQ08173.1 GL449275 EFN83057.1 KQ414851 KOC60193.1 QOIP01000005 RLU22511.1 GAKP01010680 JAC48272.1 KZ288279 PBC29675.1 KK107063 EZA60953.1 AXCM01000491 CCAG010015502 JRES01001494 KNC22482.1 ATLV01026176 KE525407 KFB52898.1 KQ434978 KZC13010.1 AAAB01008807 EAA04209.4 APCN01003702 ADMH02002070 ETN59617.1 KA648460 AFP63089.1 ACPB03010086 JXJN01023511 DS235435 EEB15722.1 GDHF01012546 JAI39768.1 CH902620 EDV31604.1 CH963920 EDW77999.1 CH954177 EDV59156.1 GDIQ01080232 JAN14505.1 GDIQ01009060 JAN85677.1 JXJN01025925 GDIQ01092428 JAL59298.1 GL732525 EFX88812.1 GDIQ01114618 JAL37108.1 LRGB01002505 KZS07282.1 GDIQ01065795 JAN28942.1 CM000157 EDW87813.1 GDIP01236719 JAI86682.1 GDIP01205460 JAJ17942.1 GDIP01235029 JAI88372.1 GDIQ01115821 JAL35905.1 CM000361 CM002910 EDX04035.1 KMY88659.1 GDIQ01048198 JAN46539.1
BABH01039390 BABH01039391 ODYU01006893 SOQ49150.1 AGBW02007696 OWR54700.1 KQ461185 KPJ07424.1 KZ149906 PZC78299.1 NWSH01000369 PCG76929.1 KQ459252 KPJ02191.1 LJIG01015976 KRT81987.1 NEVH01020337 PNF21651.1 KK852909 KDR13941.1 KQ971343 EFA04673.2 GEZM01057701 JAV72135.1 CH477298 EAT44349.1 GDUN01000889 JAN95030.1 MF433013 ATL75364.1 JXUM01150325 KQ570973 KXJ68272.1 DS232061 EDS33523.1 GFDL01009731 JAV25314.1 GFDL01009774 JAV25271.1 APGK01028388 APGK01028389 KB740686 KB631879 ENN79576.1 ERL86904.1 GEDC01005773 JAS31525.1 KQ977647 KYN00840.1 GECZ01002928 JAS66841.1 KQ979568 KYN20789.1 GEBQ01024444 JAT15533.1 KQ981864 KYN34347.1 ADTU01019317 GL888818 EGI57809.1 KQ976662 KYM78448.1 KQ982314 KYQ58004.1 GEBQ01023040 JAT16937.1 LBMM01000035 KMR05231.1 GAMC01003200 JAC03356.1 GGFL01006194 MBW70372.1 GGFL01006193 MBW70371.1 GBXI01004808 JAD09484.1 AXCN02000522 GDHC01010456 JAQ08173.1 GL449275 EFN83057.1 KQ414851 KOC60193.1 QOIP01000005 RLU22511.1 GAKP01010680 JAC48272.1 KZ288279 PBC29675.1 KK107063 EZA60953.1 AXCM01000491 CCAG010015502 JRES01001494 KNC22482.1 ATLV01026176 KE525407 KFB52898.1 KQ434978 KZC13010.1 AAAB01008807 EAA04209.4 APCN01003702 ADMH02002070 ETN59617.1 KA648460 AFP63089.1 ACPB03010086 JXJN01023511 DS235435 EEB15722.1 GDHF01012546 JAI39768.1 CH902620 EDV31604.1 CH963920 EDW77999.1 CH954177 EDV59156.1 GDIQ01080232 JAN14505.1 GDIQ01009060 JAN85677.1 JXJN01025925 GDIQ01092428 JAL59298.1 GL732525 EFX88812.1 GDIQ01114618 JAL37108.1 LRGB01002505 KZS07282.1 GDIQ01065795 JAN28942.1 CM000157 EDW87813.1 GDIP01236719 JAI86682.1 GDIP01205460 JAJ17942.1 GDIP01235029 JAI88372.1 GDIQ01115821 JAL35905.1 CM000361 CM002910 EDX04035.1 KMY88659.1 GDIQ01048198 JAN46539.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000218220
UP000053268
UP000235965
+ More
UP000027135 UP000007266 UP000008820 UP000069940 UP000249989 UP000002320 UP000019118 UP000030742 UP000078542 UP000078492 UP000078541 UP000005205 UP000007755 UP000078540 UP000075809 UP000036403 UP000069272 UP000075881 UP000075886 UP000008237 UP000053825 UP000279307 UP000242457 UP000053097 UP000075884 UP000075883 UP000078200 UP000092445 UP000092444 UP000091820 UP000076408 UP000075885 UP000037069 UP000030765 UP000075882 UP000075903 UP000076502 UP000075902 UP000007062 UP000076407 UP000075840 UP000095301 UP000075880 UP000075920 UP000000673 UP000095300 UP000015103 UP000092460 UP000009046 UP000192221 UP000007801 UP000005203 UP000075900 UP000007798 UP000008711 UP000092443 UP000000305 UP000076858 UP000002282 UP000000304
UP000027135 UP000007266 UP000008820 UP000069940 UP000249989 UP000002320 UP000019118 UP000030742 UP000078542 UP000078492 UP000078541 UP000005205 UP000007755 UP000078540 UP000075809 UP000036403 UP000069272 UP000075881 UP000075886 UP000008237 UP000053825 UP000279307 UP000242457 UP000053097 UP000075884 UP000075883 UP000078200 UP000092445 UP000092444 UP000091820 UP000076408 UP000075885 UP000037069 UP000030765 UP000075882 UP000075903 UP000076502 UP000075902 UP000007062 UP000076407 UP000075840 UP000095301 UP000075880 UP000075920 UP000000673 UP000095300 UP000015103 UP000092460 UP000009046 UP000192221 UP000007801 UP000005203 UP000075900 UP000007798 UP000008711 UP000092443 UP000000305 UP000076858 UP000002282 UP000000304
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
H9J568
A0A2H1W7V1
A0A212FLX5
A0A194QPY8
A0A2W1BTB9
A0A2A4JYP2
+ More
A0A194QFE5 A0A0T6B3F1 A0A2J7PZ83 A0A067QV78 D6WLF4 A0A1Y1LJL7 Q17D80 A0A0P6IVK3 A0A1S4F7G9 A0A291S6X8 A0A182G7H5 B0WRV8 A0A1Q3FCK6 A0A1Q3FCF5 N6UG33 A0A1B6E0R9 A0A151IH37 A0A1B6GWN1 A0A195E7H7 A0A1B6KVR2 A0A195F2E3 A0A158NLC3 F4X6M7 A0A195B1U8 A0A151XCB7 A0A1B6KZR0 A0A0J7LAK2 W8BQ30 A0A182FGC3 A0A2M4CYG0 A0A2M4CYU4 A0A182JTG5 A0A0A1XDL2 A0A182QQX9 A0A146LJ60 E2BMN9 A0A0L7QNJ8 A0A3L8DQ63 A0A034W1S0 A0A2A3ED52 A0A026WYN6 A0A182NJT9 A0A182LTU8 A0A1A9VIS1 A0A1B0A6D5 A0A1B0GAR0 A0A1A9X0J4 A0A182Y313 A0A182P135 A0A0L0BR21 A0A084WRQ4 A0A182L3H4 A0A182V4C5 A0A154PPB6 A0A182TRJ4 Q7PTA6 A0A182WWM4 A0A182I6W0 A0A1I8N2I6 A0A182JA52 A0A182WHX9 W5J562 T1PK51 A0A1I8N2I8 A0A1I8PJA4 A0A1I8PJD0 T1HNF5 A0A1I8PJ88 A0A1B0C0C7 A0A1I8PJ20 E0VQR7 A0A1W4UQH3 A0A0K8VLP5 B3MKS3 A0A088A791 A0A182R9V0 B4N064 B3N669 A0A0P6E9G5 A0A0N8ENM6 A0A1A9YKR0 A0A1B0C5B4 A0A0P5SFL2 E9FV02 A0A0P5QH58 A0A164PZ63 A0A0P6F3M4 B4NZY6 A0A0P4XU45 A0A0P5A1V2 A0A0P4Y1T3 A0A0P5R2A2 B4Q5A2 A0A0P6FHV1
A0A194QFE5 A0A0T6B3F1 A0A2J7PZ83 A0A067QV78 D6WLF4 A0A1Y1LJL7 Q17D80 A0A0P6IVK3 A0A1S4F7G9 A0A291S6X8 A0A182G7H5 B0WRV8 A0A1Q3FCK6 A0A1Q3FCF5 N6UG33 A0A1B6E0R9 A0A151IH37 A0A1B6GWN1 A0A195E7H7 A0A1B6KVR2 A0A195F2E3 A0A158NLC3 F4X6M7 A0A195B1U8 A0A151XCB7 A0A1B6KZR0 A0A0J7LAK2 W8BQ30 A0A182FGC3 A0A2M4CYG0 A0A2M4CYU4 A0A182JTG5 A0A0A1XDL2 A0A182QQX9 A0A146LJ60 E2BMN9 A0A0L7QNJ8 A0A3L8DQ63 A0A034W1S0 A0A2A3ED52 A0A026WYN6 A0A182NJT9 A0A182LTU8 A0A1A9VIS1 A0A1B0A6D5 A0A1B0GAR0 A0A1A9X0J4 A0A182Y313 A0A182P135 A0A0L0BR21 A0A084WRQ4 A0A182L3H4 A0A182V4C5 A0A154PPB6 A0A182TRJ4 Q7PTA6 A0A182WWM4 A0A182I6W0 A0A1I8N2I6 A0A182JA52 A0A182WHX9 W5J562 T1PK51 A0A1I8N2I8 A0A1I8PJA4 A0A1I8PJD0 T1HNF5 A0A1I8PJ88 A0A1B0C0C7 A0A1I8PJ20 E0VQR7 A0A1W4UQH3 A0A0K8VLP5 B3MKS3 A0A088A791 A0A182R9V0 B4N064 B3N669 A0A0P6E9G5 A0A0N8ENM6 A0A1A9YKR0 A0A1B0C5B4 A0A0P5SFL2 E9FV02 A0A0P5QH58 A0A164PZ63 A0A0P6F3M4 B4NZY6 A0A0P4XU45 A0A0P5A1V2 A0A0P4Y1T3 A0A0P5R2A2 B4Q5A2 A0A0P6FHV1
PDB
4PK7
E-value=0,
Score=2264
Ontologies
GO
PANTHER
Topology
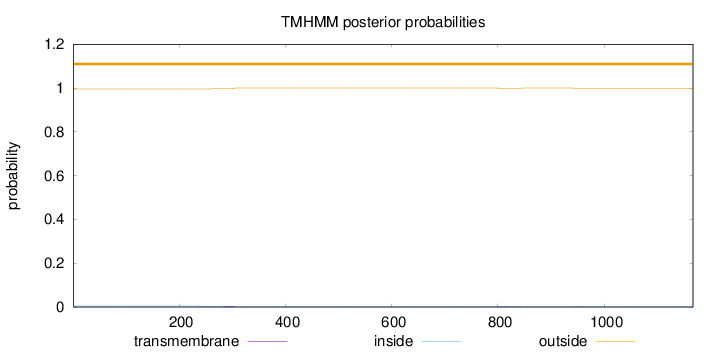
Length:
1167
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12151
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00440
outside
1 - 1167
Population Genetic Test Statistics
Pi
1.085137
Theta
5.38181
Tajima's D
-2.17155
CLR
319.132746
CSRT
0.00524973751312434
Interpretation
Possibly Positive selection
