Gene
KWMTBOMO06923
Pre Gene Modal
BGIBMGA014330
Annotation
PREDICTED:_uncharacterized_protein_LOC106125026_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 3.156
Sequence
CDS
ATGGGTAGTGAGAGTGAGAAAGTTGTAGTAGTAGTAGATGTGTTAAAGAGGAGAAGAATAAATATAGCATGTCTGCAAGAGACTAGGTGGAAGGGAGCGAAAGCTAGAGAGATTGGTGAAGGATATAAGCTGTATTATAGTGGAAGTGATGGAAGAAGGAATGGCGTGGGAGTAGTGTTAGATAAGGATTTGAAAGATTGTGTGGCGCAAGTCATAAGAACGAGTGATAGGATAATAACCGTAAAAATAGTTTGTGAAAGTGTTATTTTGAATGTTATAAGTGTGTACGCGCCTCAGTCCGAATGCAAGGAGGACGTGAAAGAAAAATTTTGGCAGGATTTTGATAGTGTTATGATTCACATTCCAGAGAGTGAAGAGATTTGTATAGGAGGTGATTTTAATGGACATGTAGGAGCTACAAATGAGGGCTATGAAAGGGTGCATGGAGGGTGGGGGTATGGCAATCGCAATGATGATGGAGACCAGTTGCTCCAAGCTGCGACCACCTTTAACCTGGCAGTGGCGAATACGTGGTTCCAGAAACGGCCTGAACACCTTATCACCTATAAGAGTGGTAACCACGCGACACAGATCGACTACTTTTTGATTAAGAGAAGTAAGTTGGTCTGTGTCAAAAACTGCAAAGTACTGCCGGGTGAAGCATTGGTGACGCAGCATAGACTGCTACTAATGGACATTACGATCAGTTACAAGTACCTAAGGAGAAAAAACCGTCTCTCGCCAAAAATTAGATGGCGTATGCTAGAGACAGATGAGTATGCTGGTCGATTTAGGGAAATAATGATAGAAAACATCTTGGAAATGAAGGATATAAAAGAGAAATCTGCGAATGATTGTTGGGATGAAATGGCAAATAGTGTCAGGAAGACAGCTAAGGCTGTGCTCGGCGAATCAAAAGGGAAAGGATACATGGTGGTGGAATGA
Protein
MGSESEKVVVVVDVLKRRRINIACLQETRWKGAKAREIGEGYKLYYSGSDGRRNGVGVVLDKDLKDCVAQVIRTSDRIITVKIVCESVILNVISVYAPQSECKEDVKEKFWQDFDSVMIHIPESEEICIGGDFNGHVGATNEGYERVHGGWGYGNRNDDGDQLLQAATTFNLAVANTWFQKRPEHLITYKSGNHATQIDYFLIKRSKLVCVKNCKVLPGEALVTQHRLLLMDITISYKYLRRKNRLSPKIRWRMLETDEYAGRFREIMIENILEMKDIKEKSANDCWDEMANSVRKTAKAVLGESKGKGYMVVE
Summary
Similarity
Belongs to the disease resistance NB-LRR family.
Uniprot
A0A2W1BQU0
A0A2A4IZ11
C6Y4D5
A0A3B1JW75
A0A2W1BJ69
A0A3L6DWK5
+ More
A0A1S3DQ07 A0A1S3XCV8 A0A1B0CS40 A0A0N4XX07 A0A2B7ZVV7 A0A1D6H2Q9 A0A0R0GIZ9 A0A3B3BNZ8 A0A3Q3RGM6 A0A3L6FAY2 A0A016VUM7 A0A2I0VYD5 A0A3L6DTP1 A0A3Q3KY92 A0A1D6MV94 A0A3L6QQ34 A0A1D6LG60 A0A3L6EGD8 A0A1S3YY63 A0A016SPI1 A0A3L6DY53 A0A3L6F033 A0A1D6FJU8 A0A1D6P7P3 A0A151SQ16 A0A1D6IXS5 A0A1D6IXS8 A0A1D6GHI2 A0A1D6PII6 A0A3L6DD55 A0A016TE69 A0A3L6FAM2 A0A1D6K3M7 G0Y6W1 A0A1D6MV72 A0A1D6J7V5 A0A1D6KWJ1 A0A1S3X0L9 A0A3P9LN16 A0A1D6PU65 A0A1D6I351 A0A1D6KX28 A0A1D6I607 A0A3L6EF28 A0A1D6KLY6 A0A1D6JQ10 A0A3L6FHK1 A0A3L6DTJ3 A0A1D6G503 A0A016TKL4 A0A1D6HX90 A0A1D6M7A6 A0A016SMN3 A0A1D6KKX4 A0A016SLX0 A0A3L6DZ28 A0A3L6G913 A0A1D6I7A1 A0A2I0BE06 A0A3L6FK19 A0A0R0JWH1 A0A1D6GYF0 A0A1D6PS19 A0A1D6J5H5 A0A0R0KL23 A0A1D6P0V1 A0A1D6NUQ3 A0A3L6GA57 A0A1D6KVT7 A0A0R0EIL6 A0A0R0LI28 A0A1D6GRX4 A0A3B3C7H7 A0A1D6NCE0 A0A016UBL1 A0A3B3E0P8 A0A1D6JJN9 A0A3B3CTW1 A0A3B3CWT9 A0A1D6LXZ4 A0A3B3CRW1 A0A3B3BQU5 A0A0R0EKU2 A0A1D6NKV0 A0A3B3B9G5 A0A3L6GCQ5 A0A1D6IVC4 A0A3B3BT53 A0A3B3CWZ8 A0A3B3CTD1 A0A3B3BH81 A0A3B3CTC7 A0A3B3BTI6 A0A3B3CQH3
A0A1S3DQ07 A0A1S3XCV8 A0A1B0CS40 A0A0N4XX07 A0A2B7ZVV7 A0A1D6H2Q9 A0A0R0GIZ9 A0A3B3BNZ8 A0A3Q3RGM6 A0A3L6FAY2 A0A016VUM7 A0A2I0VYD5 A0A3L6DTP1 A0A3Q3KY92 A0A1D6MV94 A0A3L6QQ34 A0A1D6LG60 A0A3L6EGD8 A0A1S3YY63 A0A016SPI1 A0A3L6DY53 A0A3L6F033 A0A1D6FJU8 A0A1D6P7P3 A0A151SQ16 A0A1D6IXS5 A0A1D6IXS8 A0A1D6GHI2 A0A1D6PII6 A0A3L6DD55 A0A016TE69 A0A3L6FAM2 A0A1D6K3M7 G0Y6W1 A0A1D6MV72 A0A1D6J7V5 A0A1D6KWJ1 A0A1S3X0L9 A0A3P9LN16 A0A1D6PU65 A0A1D6I351 A0A1D6KX28 A0A1D6I607 A0A3L6EF28 A0A1D6KLY6 A0A1D6JQ10 A0A3L6FHK1 A0A3L6DTJ3 A0A1D6G503 A0A016TKL4 A0A1D6HX90 A0A1D6M7A6 A0A016SMN3 A0A1D6KKX4 A0A016SLX0 A0A3L6DZ28 A0A3L6G913 A0A1D6I7A1 A0A2I0BE06 A0A3L6FK19 A0A0R0JWH1 A0A1D6GYF0 A0A1D6PS19 A0A1D6J5H5 A0A0R0KL23 A0A1D6P0V1 A0A1D6NUQ3 A0A3L6GA57 A0A1D6KVT7 A0A0R0EIL6 A0A0R0LI28 A0A1D6GRX4 A0A3B3C7H7 A0A1D6NCE0 A0A016UBL1 A0A3B3E0P8 A0A1D6JJN9 A0A3B3CTW1 A0A3B3CWT9 A0A1D6LXZ4 A0A3B3CRW1 A0A3B3BQU5 A0A0R0EKU2 A0A1D6NKV0 A0A3B3B9G5 A0A3L6GCQ5 A0A1D6IVC4 A0A3B3BT53 A0A3B3CWZ8 A0A3B3CTD1 A0A3B3BH81 A0A3B3CTC7 A0A3B3BTI6 A0A3B3CQH3
Pubmed
EMBL
KZ150068
PZC74093.1
NWSH01004369
PCG65177.1
CU462842
CBA11992.1
+ More
KZ150020 PZC74908.1 NCVQ01000008 PWZ13005.1 AJWK01025609 AJWK01025610 AJWK01025611 UYSL01019892 VDL71065.1 PDHN01001019 PGH37420.1 CM000781 AQK69123.1 CM000847 KRH15858.1 NCVQ01000004 PWZ30100.1 JARK01001340 EYC31309.1 KZ503093 PKU68428.1 NCVQ01000009 PWZ11227.1 CM007649 ONM32741.1 PQIB02000011 RLM84825.1 CM000782 AQK78905.1 NCVQ01000006 PWZ20102.1 JARK01001532 EYB92234.1 PWZ13495.1 NCVQ01000005 PWZ26506.1 CM000784 AQK92036.1 CM000785 AQL05851.1 CM003613 KYP56875.1 CM000786 AQK40715.1 AQK40710.1 AQK62958.1 AQL09133.1 NCVQ01000010 PWZ06043.1 JARK01001448 EYC00913.1 PWZ29990.1 CM000780 CM007647 AQK53188.1 ONL98233.1 HQ637178 AEL30371.1 ONM32720.1 AQK44003.1 ONM06836.1 AQK50169.1 CM007650 ONM54584.1 ONM07016.1 ONM55524.1 NCVQ01000007 PWZ18651.1 ONM03863.1 AQK59578.1 ONL94081.1 ONM09892.1 ONM36701.1 PWZ32579.1 PWZ11925.1 AQK98364.1 JARK01001431 JARK01001389 JARK01001372 EYC03231.1 EYC10881.1 EYC15468.1 ONM52836.1 AQK86945.1 JARK01001540 EYB91592.1 ONM03552.1 EYB91590.1 PWZ13333.1 NCVQ01000002 PWZ44991.1 ONM55942.1 KZ451888 PKA66012.1 PWZ33556.1 CM000839 KRH54927.1 AQK67802.1 AQK49513.1 AQK43188.1 CM000837 KRH63599.1 AQL03708.1 AQL01871.1 PWZ45158.1 ONM06616.1 CM000853 KRG89944.1 CM000834 KRH75631.1 AQK65817.1 ONM38170.1 JARK01001384 EYC12222.1 ONL92524.1 AQK84025.1 KRG90881.1 ONM40856.1 PWZ46302.1 AQK39954.1
KZ150020 PZC74908.1 NCVQ01000008 PWZ13005.1 AJWK01025609 AJWK01025610 AJWK01025611 UYSL01019892 VDL71065.1 PDHN01001019 PGH37420.1 CM000781 AQK69123.1 CM000847 KRH15858.1 NCVQ01000004 PWZ30100.1 JARK01001340 EYC31309.1 KZ503093 PKU68428.1 NCVQ01000009 PWZ11227.1 CM007649 ONM32741.1 PQIB02000011 RLM84825.1 CM000782 AQK78905.1 NCVQ01000006 PWZ20102.1 JARK01001532 EYB92234.1 PWZ13495.1 NCVQ01000005 PWZ26506.1 CM000784 AQK92036.1 CM000785 AQL05851.1 CM003613 KYP56875.1 CM000786 AQK40715.1 AQK40710.1 AQK62958.1 AQL09133.1 NCVQ01000010 PWZ06043.1 JARK01001448 EYC00913.1 PWZ29990.1 CM000780 CM007647 AQK53188.1 ONL98233.1 HQ637178 AEL30371.1 ONM32720.1 AQK44003.1 ONM06836.1 AQK50169.1 CM007650 ONM54584.1 ONM07016.1 ONM55524.1 NCVQ01000007 PWZ18651.1 ONM03863.1 AQK59578.1 ONL94081.1 ONM09892.1 ONM36701.1 PWZ32579.1 PWZ11925.1 AQK98364.1 JARK01001431 JARK01001389 JARK01001372 EYC03231.1 EYC10881.1 EYC15468.1 ONM52836.1 AQK86945.1 JARK01001540 EYB91592.1 ONM03552.1 EYB91590.1 PWZ13333.1 NCVQ01000002 PWZ44991.1 ONM55942.1 KZ451888 PKA66012.1 PWZ33556.1 CM000839 KRH54927.1 AQK67802.1 AQK49513.1 AQK43188.1 CM000837 KRH63599.1 AQL03708.1 AQL01871.1 PWZ45158.1 ONM06616.1 CM000853 KRG89944.1 CM000834 KRH75631.1 AQK65817.1 ONM38170.1 JARK01001384 EYC12222.1 ONL92524.1 AQK84025.1 KRG90881.1 ONM40856.1 PWZ46302.1 AQK39954.1
Proteomes
Pfam
Interpro
IPR036691
Endo/exonu/phosph_ase_sf
+ More
IPR027124 Swc5/CFDP2
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR036877 SUI1_dom_sf
IPR036885 SWIB_MDM2_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR025585 PSII_Pbs27
IPR038450 PSII_Pbs27_sf
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR017441 Protein_kinase_ATP_BS
IPR032675 LRR_dom_sf
IPR027417 P-loop_NTPase
IPR002182 NB-ARC
IPR036390 WH_DNA-bd_sf
IPR036865 CRAL-TRIO_dom_sf
IPR036273 CRAL/TRIO_N_dom_sf
IPR036291 NAD(P)-bd_dom_sf
IPR002347 SDR_fam
IPR013865 FAM32A
IPR027124 Swc5/CFDP2
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR036877 SUI1_dom_sf
IPR036885 SWIB_MDM2_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR025585 PSII_Pbs27
IPR038450 PSII_Pbs27_sf
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR017441 Protein_kinase_ATP_BS
IPR032675 LRR_dom_sf
IPR027417 P-loop_NTPase
IPR002182 NB-ARC
IPR036390 WH_DNA-bd_sf
IPR036865 CRAL-TRIO_dom_sf
IPR036273 CRAL/TRIO_N_dom_sf
IPR036291 NAD(P)-bd_dom_sf
IPR002347 SDR_fam
IPR013865 FAM32A
SUPFAM
ProteinModelPortal
A0A2W1BQU0
A0A2A4IZ11
C6Y4D5
A0A3B1JW75
A0A2W1BJ69
A0A3L6DWK5
+ More
A0A1S3DQ07 A0A1S3XCV8 A0A1B0CS40 A0A0N4XX07 A0A2B7ZVV7 A0A1D6H2Q9 A0A0R0GIZ9 A0A3B3BNZ8 A0A3Q3RGM6 A0A3L6FAY2 A0A016VUM7 A0A2I0VYD5 A0A3L6DTP1 A0A3Q3KY92 A0A1D6MV94 A0A3L6QQ34 A0A1D6LG60 A0A3L6EGD8 A0A1S3YY63 A0A016SPI1 A0A3L6DY53 A0A3L6F033 A0A1D6FJU8 A0A1D6P7P3 A0A151SQ16 A0A1D6IXS5 A0A1D6IXS8 A0A1D6GHI2 A0A1D6PII6 A0A3L6DD55 A0A016TE69 A0A3L6FAM2 A0A1D6K3M7 G0Y6W1 A0A1D6MV72 A0A1D6J7V5 A0A1D6KWJ1 A0A1S3X0L9 A0A3P9LN16 A0A1D6PU65 A0A1D6I351 A0A1D6KX28 A0A1D6I607 A0A3L6EF28 A0A1D6KLY6 A0A1D6JQ10 A0A3L6FHK1 A0A3L6DTJ3 A0A1D6G503 A0A016TKL4 A0A1D6HX90 A0A1D6M7A6 A0A016SMN3 A0A1D6KKX4 A0A016SLX0 A0A3L6DZ28 A0A3L6G913 A0A1D6I7A1 A0A2I0BE06 A0A3L6FK19 A0A0R0JWH1 A0A1D6GYF0 A0A1D6PS19 A0A1D6J5H5 A0A0R0KL23 A0A1D6P0V1 A0A1D6NUQ3 A0A3L6GA57 A0A1D6KVT7 A0A0R0EIL6 A0A0R0LI28 A0A1D6GRX4 A0A3B3C7H7 A0A1D6NCE0 A0A016UBL1 A0A3B3E0P8 A0A1D6JJN9 A0A3B3CTW1 A0A3B3CWT9 A0A1D6LXZ4 A0A3B3CRW1 A0A3B3BQU5 A0A0R0EKU2 A0A1D6NKV0 A0A3B3B9G5 A0A3L6GCQ5 A0A1D6IVC4 A0A3B3BT53 A0A3B3CWZ8 A0A3B3CTD1 A0A3B3BH81 A0A3B3CTC7 A0A3B3BTI6 A0A3B3CQH3
A0A1S3DQ07 A0A1S3XCV8 A0A1B0CS40 A0A0N4XX07 A0A2B7ZVV7 A0A1D6H2Q9 A0A0R0GIZ9 A0A3B3BNZ8 A0A3Q3RGM6 A0A3L6FAY2 A0A016VUM7 A0A2I0VYD5 A0A3L6DTP1 A0A3Q3KY92 A0A1D6MV94 A0A3L6QQ34 A0A1D6LG60 A0A3L6EGD8 A0A1S3YY63 A0A016SPI1 A0A3L6DY53 A0A3L6F033 A0A1D6FJU8 A0A1D6P7P3 A0A151SQ16 A0A1D6IXS5 A0A1D6IXS8 A0A1D6GHI2 A0A1D6PII6 A0A3L6DD55 A0A016TE69 A0A3L6FAM2 A0A1D6K3M7 G0Y6W1 A0A1D6MV72 A0A1D6J7V5 A0A1D6KWJ1 A0A1S3X0L9 A0A3P9LN16 A0A1D6PU65 A0A1D6I351 A0A1D6KX28 A0A1D6I607 A0A3L6EF28 A0A1D6KLY6 A0A1D6JQ10 A0A3L6FHK1 A0A3L6DTJ3 A0A1D6G503 A0A016TKL4 A0A1D6HX90 A0A1D6M7A6 A0A016SMN3 A0A1D6KKX4 A0A016SLX0 A0A3L6DZ28 A0A3L6G913 A0A1D6I7A1 A0A2I0BE06 A0A3L6FK19 A0A0R0JWH1 A0A1D6GYF0 A0A1D6PS19 A0A1D6J5H5 A0A0R0KL23 A0A1D6P0V1 A0A1D6NUQ3 A0A3L6GA57 A0A1D6KVT7 A0A0R0EIL6 A0A0R0LI28 A0A1D6GRX4 A0A3B3C7H7 A0A1D6NCE0 A0A016UBL1 A0A3B3E0P8 A0A1D6JJN9 A0A3B3CTW1 A0A3B3CWT9 A0A1D6LXZ4 A0A3B3CRW1 A0A3B3BQU5 A0A0R0EKU2 A0A1D6NKV0 A0A3B3B9G5 A0A3L6GCQ5 A0A1D6IVC4 A0A3B3BT53 A0A3B3CWZ8 A0A3B3CTD1 A0A3B3BH81 A0A3B3CTC7 A0A3B3BTI6 A0A3B3CQH3
PDB
2VOA
E-value=0.0132718,
Score=89
Ontologies
PANTHER
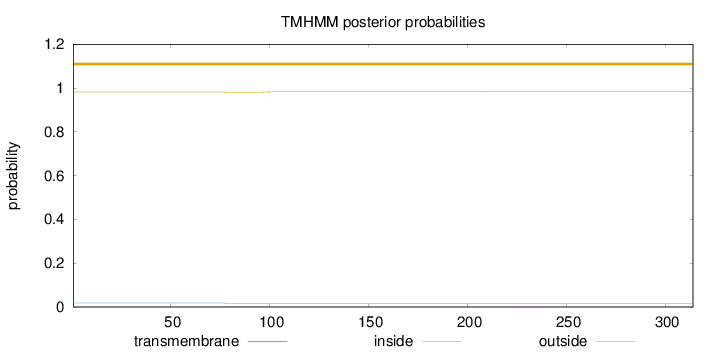
Topology
Length:
314
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04589
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01861
outside
1 - 314
Population Genetic Test Statistics
Pi
17.894648
Theta
18.596138
Tajima's D
-0.599241
CLR
4.169753
CSRT
0.222738863056847
Interpretation
Uncertain
