Gene
KWMTBOMO06922
Pre Gene Modal
BGIBMGA004639
Annotation
PREDICTED:_frizzled-2-like_[Bombyx_mori]
Full name
Frizzled-8
+ More
Frizzled-5
Frizzled-5
Location in the cell
PlasmaMembrane Reliability : 3.524
Sequence
CDS
ATGTGGGCAGCGGGCGTGGCGGCGTGCGCGCTGCTGGCCGCCGCCGGAGCCCTGCAGCAGCCGCGCTGTCAGGACATCACCATCCCCATGTGCCGCGGCATCGGCTACAACCTCACTTCCTTCCCCAACGCGCTGGACCACGACACGCAGGAAGAAGCCGGACTAGAGGTTCACCAATATTGGCCCCTCGTCGAGATCAAGTGCTCGTCGGACCTAAAGTTCTTCCTGTGCTCCGTGTACACGCCGATCTGCATCGAGGACTACGCGAAGCCGCTGCCGGCGTGTCGCAGCGTGTGCGAGCGCGCGCGGGCCGGCTGCGCGCCGCTCATGCAGCAGTACGGGTTCCCGTGGCCCGAGCGCATGGCGTGCGAGCAGCTGCCGCGCGCCGGCGACCCCGACCAGCTGTGCATGGAGGAGACGGAGCGCGCGCACGAGCCCGAGCCGCCCCGCCCGCCGCCCCGCAAGCCCGCCAAGAACAACTGCAAGGATCCAAAAAACTGCGGCGGCGCGGCGGCTGCGGGCGCCGGCGACGCGGCCGAGTGCGAGTGCGCGTGCCGCCCGCCGCTGCTCGCCGCGCCCAACACCAGCGCCGCGCCCGGCCTGCGCCCCCCCGCCTGTCTGCAGCCCTGCCGCGGCGCCTTCTTCACCGCCGACGAAAAACACTTCGCGGCCGTCTGGGTGGCGCTGTGGAGCGGCCTCTGCGCCGCCTCCACGCTCATGACGCTCACCACCTTCCTGATCGACTCGCAGCGGTTTAAGTACCCGGAGCGCCCCATCGTGTACTTGTCGGCCTGCTACTTCATGGTGTCGCTGGGCTACCTGGCGCGCCTGGCGCTGGGGCACGACGAGATCGCGTGCGACGGGCCGGCGCTCAAGGTGTCCGCCAGCGGACCCAGCGCGTGCACGCTCGTCTTCATCTTGGTCTATTTCTTCGGAATGGCGTCGTCGATCTGGTGGGTGGTGCTGTCCTTCGCGTGGTTCCTCGCCGCCGGACTCAAGTGGGGCAATGAAGCCATCGCCGGTTACGCACAGTACTACCACCTGGCCGCGTGGCTCGTACCGGCTGCGAAGACGGTCGCCGTGCTGCTGGCCGGCGCCGTTGACGGCGACCCGGTGGCCGGGATCTGTTCAGTCGGCAACTCTTCTCCGGAAAATCTCAAGAAATTCGTGCTGGCGCCGCTGGTCGTGTACTTCGGTCTGGGCGCGACGTTCCTGCTCGCCGGGTTCGTGTCGCTGTTCCGCATCCGGTCGGTGATCAAGCGGCAGGGCGGCGTGGGCGCGGGCTCCAAGGCGGACAAGCTGGAGAAGCTGATGATCCGCATCGGCGTGTTCAGCGTGCTGTACGCGGTGCCGGCGGGCGCGGTGCTGGCGTGCCTGGCGTACGAGGCGGCGGGGCACGCGGGCTGGCTGCGGCGCGCCGCGTGCGGCTCCCGCTGCGGGCCGGCGCCGCTGTACGCCGCGCTCATGCTCAAGTACTTCATGGCGCTGGCGGTGGGCATCACGTCGGGCGTGTGGATCTGGTCGGGCAAGACGCTGGACTCGTGGCGCCGCGTGTGGGCGGGCGGGCGGCGCGTGCCCCCGCCCTCGCAGCGCGCGCTCGTCAAGGGCGCCGTGTGA
Protein
MWAAGVAACALLAAAGALQQPRCQDITIPMCRGIGYNLTSFPNALDHDTQEEAGLEVHQYWPLVEIKCSSDLKFFLCSVYTPICIEDYAKPLPACRSVCERARAGCAPLMQQYGFPWPERMACEQLPRAGDPDQLCMEETERAHEPEPPRPPPRKPAKNNCKDPKNCGGAAAAGAGDAAECECACRPPLLAAPNTSAAPGLRPPACLQPCRGAFFTADEKHFAAVWVALWSGLCAASTLMTLTTFLIDSQRFKYPERPIVYLSACYFMVSLGYLARLALGHDEIACDGPALKVSASGPSACTLVFILVYFFGMASSIWWVVLSFAWFLAAGLKWGNEAIAGYAQYYHLAAWLVPAAKTVAVLLAGAVDGDPVAGICSVGNSSPENLKKFVLAPLVVYFGLGATFLLAGFVSLFRIRSVIKRQGGVGAGSKADKLEKLMIRIGVFSVLYAVPAGAVLACLAYEAAGHAGWLRRAACGSRCGPAPLYAALMLKYFMALAVGITSGVWIWSGKTLDSWRRVWAGGRRVPPPSQRALVKGAV
Summary
Description
Receptor for Wnt proteins. Most of frizzled receptors are coupled to the beta-catenin canonical signaling pathway, which leads to the activation of disheveled proteins, inhibition of GSK-3 kinase, nuclear accumulation of beta-catenin and activation of Wnt target genes. A second signaling pathway involving PKC and calcium fluxes has been seen for some family members, but it is not yet clear if it represents a distinct pathway or if it can be integrated in the canonical pathway, as PKC seems to be required for Wnt-mediated inactivation of GSK-3 kinase. Both pathways seem to involve interactions with G-proteins. May be involved in transduction and intercellular transmission of polarity information during tissue morphogenesis and/or in differentiated tissues. Activation by Wnt8, Wnt5A or Wnt3A induces expression of beta-catenin target genes. Displays an axis-inducing activity.
Receptor for Wnt proteins. Functions in the canonical Wnt/beta-catenin signaling pathway. The canonical Wnt/beta-catenin signaling pathway leads to the activation of disheveled proteins, inhibition of GSK-3 kinase, nuclear accumulation of beta-catenin and activation of Wnt target genes (By similarity). A second signaling pathway involving PKC and calcium fluxes has been seen for some family members, but it is not yet clear if it represents a distinct pathway or if it can be integrated in the canonical pathway, as PKC seems to be required for Wnt-mediated inactivation of GSK-3 kinase. Both pathways seem to involve interactions with G-proteins. May be involved in transduction and intercellular transmission of polarity information during tissue morphogenesis and/or in differentiated tissues (Probable).
Receptor for Wnt proteins. Functions in the canonical Wnt/beta-catenin signaling pathway. The canonical Wnt/beta-catenin signaling pathway leads to the activation of disheveled proteins, inhibition of GSK-3 kinase, nuclear accumulation of beta-catenin and activation of Wnt target genes (By similarity). A second signaling pathway involving PKC and calcium fluxes has been seen for some family members, but it is not yet clear if it represents a distinct pathway or if it can be integrated in the canonical pathway, as PKC seems to be required for Wnt-mediated inactivation of GSK-3 kinase. Both pathways seem to involve interactions with G-proteins. May be involved in transduction and intercellular transmission of polarity information during tissue morphogenesis and/or in differentiated tissues (Probable).
Subunit
Interacts with lypd6 and the interaction is strongly enhanced by wnt3a (PubMed:23987510).
Similarity
Belongs to the G-protein coupled receptor Fz/Smo family.
Keywords
Cell membrane
Developmental protein
Disulfide bond
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Signal
Transducer
Transmembrane
Transmembrane helix
Wnt signaling pathway
Golgi apparatus
Feature
chain Frizzled-8
Uniprot
A0A212FLT4
A0A2H1V370
A0A1L8DK48
A0A182H9U6
A0A1S4FER6
Q174W8
+ More
B0XF00 A0A1B0D459 A0A182YIP9 D6WG76 A0A2P8XU91 A0A0A9X185 A0A1B6C5G0 T1HKP7 U4URZ2 N6T4T2 A0A0L7QT69 A0A1Q3F425 A0A1Q3F4C5 A0A1Q3F402 A0A2J7R9R2 A0A224X9S1 A0A336MTN4 A0A336LSY3 A0A1Y1K6Z8 K7JBY1 A0A2R7WR03 A0A154PC89 A0A1A9X0F7 A0A067RPY7 E0VGL1 A0A143VP63 A0A1S3KHK3 A0A193PMF2 A0A147A2P6 A0A194Q9T8 A0A3N0Z9R6 A0A3B4D8K0 H3B9V4 A0A2D0R765 A0A3P8VZI9 A0A2P4SHJ0 A0A0F8C7W7 A0A1L8EX74 A0A3Q2VUY7 A0A3P9N8F6 W5LUP7 B3DIG4 D1LX20 W4Z465 Q9W6E2 A0A1L8FVD0 A0A087YPZ1 A0A1V4L046 F6QAW3 O93274 A0A3Q2HVX2 A0A3B3X1I2 G3NBP6 A0A3B3TKC7 A0A1A8CC31 A0A3P8RSQ2 Q2WC00 A0A2I4DCZ9 A0A3Q1CWG3 A0A3Q3SQ36 A0A3B4B9X8 A0A3Q3G2M5 A0A3B4YN81 W5U7U1 A0A2S2P2Z4 M3ZGK7 A0A3Q4G1N3 A0A3Q1HHE2 A0A3B4U8Y5 A0A2U9CCF7 A0A2I0LXL0 A0A146P5Q0 A0A3P8UTE2 A0A3B3SLN2 A0A0Q3M2V1 B7ZQV9 A0A287BDT2 A0A3B4FT02 A0A3Q1FTC3 A0A3P8QQD2 A0A3P9CS68 A0A3Q2ECT1 A0A2H8TH98 A0A3B5BG11 F6VM33 B7ZPB7 P58421 A0A1L8EPX2 A0A3B5MXP3 A0A3L8Q5P4 A0A3B4EPW2 F7AS02 A0A3Q3EVM8
B0XF00 A0A1B0D459 A0A182YIP9 D6WG76 A0A2P8XU91 A0A0A9X185 A0A1B6C5G0 T1HKP7 U4URZ2 N6T4T2 A0A0L7QT69 A0A1Q3F425 A0A1Q3F4C5 A0A1Q3F402 A0A2J7R9R2 A0A224X9S1 A0A336MTN4 A0A336LSY3 A0A1Y1K6Z8 K7JBY1 A0A2R7WR03 A0A154PC89 A0A1A9X0F7 A0A067RPY7 E0VGL1 A0A143VP63 A0A1S3KHK3 A0A193PMF2 A0A147A2P6 A0A194Q9T8 A0A3N0Z9R6 A0A3B4D8K0 H3B9V4 A0A2D0R765 A0A3P8VZI9 A0A2P4SHJ0 A0A0F8C7W7 A0A1L8EX74 A0A3Q2VUY7 A0A3P9N8F6 W5LUP7 B3DIG4 D1LX20 W4Z465 Q9W6E2 A0A1L8FVD0 A0A087YPZ1 A0A1V4L046 F6QAW3 O93274 A0A3Q2HVX2 A0A3B3X1I2 G3NBP6 A0A3B3TKC7 A0A1A8CC31 A0A3P8RSQ2 Q2WC00 A0A2I4DCZ9 A0A3Q1CWG3 A0A3Q3SQ36 A0A3B4B9X8 A0A3Q3G2M5 A0A3B4YN81 W5U7U1 A0A2S2P2Z4 M3ZGK7 A0A3Q4G1N3 A0A3Q1HHE2 A0A3B4U8Y5 A0A2U9CCF7 A0A2I0LXL0 A0A146P5Q0 A0A3P8UTE2 A0A3B3SLN2 A0A0Q3M2V1 B7ZQV9 A0A287BDT2 A0A3B4FT02 A0A3Q1FTC3 A0A3P8QQD2 A0A3P9CS68 A0A3Q2ECT1 A0A2H8TH98 A0A3B5BG11 F6VM33 B7ZPB7 P58421 A0A1L8EPX2 A0A3B5MXP3 A0A3L8Q5P4 A0A3B4EPW2 F7AS02 A0A3Q3EVM8
Pubmed
22118469
26483478
17510324
25244985
18362917
19820115
+ More
29403074 25401762 26823975 23537049 28004739 20075255 24845553 20566863 27346542 26354079 9215903 24487278 25835551 27762356 25329095 23594743 20431018 9651509 9636083 10097073 23987510 19892987 16722215 16396908 25463417 23127152 23542700 25186727 23371554 29240929 11335120 30282656
29403074 25401762 26823975 23537049 28004739 20075255 24845553 20566863 27346542 26354079 9215903 24487278 25835551 27762356 25329095 23594743 20431018 9651509 9636083 10097073 23987510 19892987 16722215 16396908 25463417 23127152 23542700 25186727 23371554 29240929 11335120 30282656
EMBL
AGBW02007696
OWR54705.1
ODYU01000452
SOQ35277.1
GFDF01007337
JAV06747.1
+ More
JXUM01004841 KQ560182 KXJ83965.1 CH477405 EAT41596.2 DS232885 EDS26348.1 AJVK01003115 KQ971320 EFA01325.1 PYGN01001337 PSN35572.1 GBHO01032739 GDHC01013891 JAG10865.1 JAQ04738.1 GEDC01028823 GEDC01020202 JAS08475.1 JAS17096.1 ACPB03015808 KB632338 ERL92926.1 APGK01043959 KB741019 ENN75149.1 KQ414755 KOC61754.1 GFDL01012723 JAV22322.1 GFDL01012707 JAV22338.1 GFDL01012764 JAV22281.1 NEVH01006580 PNF37566.1 GFTR01007340 JAW09086.1 UFQS01002678 UFQT01002678 SSX14523.1 SSX33924.1 UFQT01000098 SSX19833.1 GEZM01093585 GEZM01093584 JAV56238.1 AAZX01009892 KK855322 PTY22006.1 KQ434870 KZC09539.1 KK852538 KDR21799.1 DS235149 EEB12517.1 LN849083 CRI73785.1 LC138024 BAV17685.1 GCES01013535 JAR72788.1 KQ459252 KPJ02184.1 RJVU01002418 ROL55062.1 AFYH01069935 PPHD01048218 POI23543.1 KQ042628 KKF12960.1 CM004482 OCT63879.1 FQ377896 BC163118 BC163120 AAI63118.1 GU075997 ACY92526.1 AAGJ04009227 AF117387 AAD25357.1 CM004476 OCT75534.1 AYCK01002502 LSYS01000429 OPJ90213.1 AAMC01066237 AF017177 AF033110 HADZ01012364 SBP76305.1 AM084899 CAJ29888.1 JT407661 AHH38066.1 GGMR01011218 MBY23837.1 CP026256 AWP13456.1 AKCR02000066 PKK22172.1 GCES01147227 JAQ39095.1 LMAW01002801 KQK76912.1 BC169947 BC169949 CM004477 AAI69947.1 OCT73890.1 AEMK02000074 GFXV01001688 MBW13493.1 AAMC01042393 BC169396 BC169398 AAI69396.1 AF300716 CM004483 OCT61360.1 QUSF01006046 RLV62647.1
JXUM01004841 KQ560182 KXJ83965.1 CH477405 EAT41596.2 DS232885 EDS26348.1 AJVK01003115 KQ971320 EFA01325.1 PYGN01001337 PSN35572.1 GBHO01032739 GDHC01013891 JAG10865.1 JAQ04738.1 GEDC01028823 GEDC01020202 JAS08475.1 JAS17096.1 ACPB03015808 KB632338 ERL92926.1 APGK01043959 KB741019 ENN75149.1 KQ414755 KOC61754.1 GFDL01012723 JAV22322.1 GFDL01012707 JAV22338.1 GFDL01012764 JAV22281.1 NEVH01006580 PNF37566.1 GFTR01007340 JAW09086.1 UFQS01002678 UFQT01002678 SSX14523.1 SSX33924.1 UFQT01000098 SSX19833.1 GEZM01093585 GEZM01093584 JAV56238.1 AAZX01009892 KK855322 PTY22006.1 KQ434870 KZC09539.1 KK852538 KDR21799.1 DS235149 EEB12517.1 LN849083 CRI73785.1 LC138024 BAV17685.1 GCES01013535 JAR72788.1 KQ459252 KPJ02184.1 RJVU01002418 ROL55062.1 AFYH01069935 PPHD01048218 POI23543.1 KQ042628 KKF12960.1 CM004482 OCT63879.1 FQ377896 BC163118 BC163120 AAI63118.1 GU075997 ACY92526.1 AAGJ04009227 AF117387 AAD25357.1 CM004476 OCT75534.1 AYCK01002502 LSYS01000429 OPJ90213.1 AAMC01066237 AF017177 AF033110 HADZ01012364 SBP76305.1 AM084899 CAJ29888.1 JT407661 AHH38066.1 GGMR01011218 MBY23837.1 CP026256 AWP13456.1 AKCR02000066 PKK22172.1 GCES01147227 JAQ39095.1 LMAW01002801 KQK76912.1 BC169947 BC169949 CM004477 AAI69947.1 OCT73890.1 AEMK02000074 GFXV01001688 MBW13493.1 AAMC01042393 BC169396 BC169398 AAI69396.1 AF300716 CM004483 OCT61360.1 QUSF01006046 RLV62647.1
Proteomes
UP000007151
UP000069940
UP000249989
UP000008820
UP000002320
UP000092462
+ More
UP000076408 UP000007266 UP000245037 UP000015103 UP000030742 UP000019118 UP000053825 UP000235965 UP000002358 UP000076502 UP000091820 UP000027135 UP000009046 UP000085678 UP000053268 UP000261440 UP000008672 UP000221080 UP000265120 UP000186698 UP000264840 UP000242638 UP000018467 UP000000437 UP000007110 UP000028760 UP000190648 UP000008143 UP000002281 UP000261480 UP000007635 UP000261500 UP000265080 UP000192220 UP000257160 UP000261640 UP000261520 UP000261660 UP000261360 UP000002852 UP000261580 UP000265040 UP000261420 UP000246464 UP000053872 UP000265000 UP000261540 UP000051836 UP000008227 UP000261460 UP000257200 UP000265100 UP000265160 UP000265020 UP000261400 UP000261380 UP000276834 UP000264800
UP000076408 UP000007266 UP000245037 UP000015103 UP000030742 UP000019118 UP000053825 UP000235965 UP000002358 UP000076502 UP000091820 UP000027135 UP000009046 UP000085678 UP000053268 UP000261440 UP000008672 UP000221080 UP000265120 UP000186698 UP000264840 UP000242638 UP000018467 UP000000437 UP000007110 UP000028760 UP000190648 UP000008143 UP000002281 UP000261480 UP000007635 UP000261500 UP000265080 UP000192220 UP000257160 UP000261640 UP000261520 UP000261660 UP000261360 UP000002852 UP000261580 UP000265040 UP000261420 UP000246464 UP000053872 UP000265000 UP000261540 UP000051836 UP000008227 UP000261460 UP000257200 UP000265100 UP000265160 UP000265020 UP000261400 UP000261380 UP000276834 UP000264800
PRIDE
Interpro
SUPFAM
SSF63501
SSF63501
Gene 3D
ProteinModelPortal
A0A212FLT4
A0A2H1V370
A0A1L8DK48
A0A182H9U6
A0A1S4FER6
Q174W8
+ More
B0XF00 A0A1B0D459 A0A182YIP9 D6WG76 A0A2P8XU91 A0A0A9X185 A0A1B6C5G0 T1HKP7 U4URZ2 N6T4T2 A0A0L7QT69 A0A1Q3F425 A0A1Q3F4C5 A0A1Q3F402 A0A2J7R9R2 A0A224X9S1 A0A336MTN4 A0A336LSY3 A0A1Y1K6Z8 K7JBY1 A0A2R7WR03 A0A154PC89 A0A1A9X0F7 A0A067RPY7 E0VGL1 A0A143VP63 A0A1S3KHK3 A0A193PMF2 A0A147A2P6 A0A194Q9T8 A0A3N0Z9R6 A0A3B4D8K0 H3B9V4 A0A2D0R765 A0A3P8VZI9 A0A2P4SHJ0 A0A0F8C7W7 A0A1L8EX74 A0A3Q2VUY7 A0A3P9N8F6 W5LUP7 B3DIG4 D1LX20 W4Z465 Q9W6E2 A0A1L8FVD0 A0A087YPZ1 A0A1V4L046 F6QAW3 O93274 A0A3Q2HVX2 A0A3B3X1I2 G3NBP6 A0A3B3TKC7 A0A1A8CC31 A0A3P8RSQ2 Q2WC00 A0A2I4DCZ9 A0A3Q1CWG3 A0A3Q3SQ36 A0A3B4B9X8 A0A3Q3G2M5 A0A3B4YN81 W5U7U1 A0A2S2P2Z4 M3ZGK7 A0A3Q4G1N3 A0A3Q1HHE2 A0A3B4U8Y5 A0A2U9CCF7 A0A2I0LXL0 A0A146P5Q0 A0A3P8UTE2 A0A3B3SLN2 A0A0Q3M2V1 B7ZQV9 A0A287BDT2 A0A3B4FT02 A0A3Q1FTC3 A0A3P8QQD2 A0A3P9CS68 A0A3Q2ECT1 A0A2H8TH98 A0A3B5BG11 F6VM33 B7ZPB7 P58421 A0A1L8EPX2 A0A3B5MXP3 A0A3L8Q5P4 A0A3B4EPW2 F7AS02 A0A3Q3EVM8
B0XF00 A0A1B0D459 A0A182YIP9 D6WG76 A0A2P8XU91 A0A0A9X185 A0A1B6C5G0 T1HKP7 U4URZ2 N6T4T2 A0A0L7QT69 A0A1Q3F425 A0A1Q3F4C5 A0A1Q3F402 A0A2J7R9R2 A0A224X9S1 A0A336MTN4 A0A336LSY3 A0A1Y1K6Z8 K7JBY1 A0A2R7WR03 A0A154PC89 A0A1A9X0F7 A0A067RPY7 E0VGL1 A0A143VP63 A0A1S3KHK3 A0A193PMF2 A0A147A2P6 A0A194Q9T8 A0A3N0Z9R6 A0A3B4D8K0 H3B9V4 A0A2D0R765 A0A3P8VZI9 A0A2P4SHJ0 A0A0F8C7W7 A0A1L8EX74 A0A3Q2VUY7 A0A3P9N8F6 W5LUP7 B3DIG4 D1LX20 W4Z465 Q9W6E2 A0A1L8FVD0 A0A087YPZ1 A0A1V4L046 F6QAW3 O93274 A0A3Q2HVX2 A0A3B3X1I2 G3NBP6 A0A3B3TKC7 A0A1A8CC31 A0A3P8RSQ2 Q2WC00 A0A2I4DCZ9 A0A3Q1CWG3 A0A3Q3SQ36 A0A3B4B9X8 A0A3Q3G2M5 A0A3B4YN81 W5U7U1 A0A2S2P2Z4 M3ZGK7 A0A3Q4G1N3 A0A3Q1HHE2 A0A3B4U8Y5 A0A2U9CCF7 A0A2I0LXL0 A0A146P5Q0 A0A3P8UTE2 A0A3B3SLN2 A0A0Q3M2V1 B7ZQV9 A0A287BDT2 A0A3B4FT02 A0A3Q1FTC3 A0A3P8QQD2 A0A3P9CS68 A0A3Q2ECT1 A0A2H8TH98 A0A3B5BG11 F6VM33 B7ZPB7 P58421 A0A1L8EPX2 A0A3B5MXP3 A0A3L8Q5P4 A0A3B4EPW2 F7AS02 A0A3Q3EVM8
PDB
6BD4
E-value=4.15667e-80,
Score=760
Ontologies
PATHWAY
GO
GO:0007275
GO:0016021
GO:0004888
GO:0016055
GO:0007166
GO:0005886
GO:0035567
GO:0017147
GO:0042813
GO:0060070
GO:0090090
GO:0043010
GO:0048794
GO:0001889
GO:0051091
GO:0004930
GO:0045595
GO:0000139
GO:0042127
GO:0048666
GO:0021531
GO:0060898
GO:0021879
GO:0021529
GO:0016020
GO:0005515
GO:0043565
GO:0043401
GO:0006418
GO:0005634
GO:0006260
PANTHER
Topology
Subcellular location
Membrane
Cell membrane
Golgi apparatus membrane Localized at the plasma membrane and also found at the Golgi. With evidence from 2 publications.
Cell membrane
Golgi apparatus membrane Localized at the plasma membrane and also found at the Golgi. With evidence from 2 publications.
SignalP
Position: 1 - 17,
Likelihood: 0.982823

Length:
538
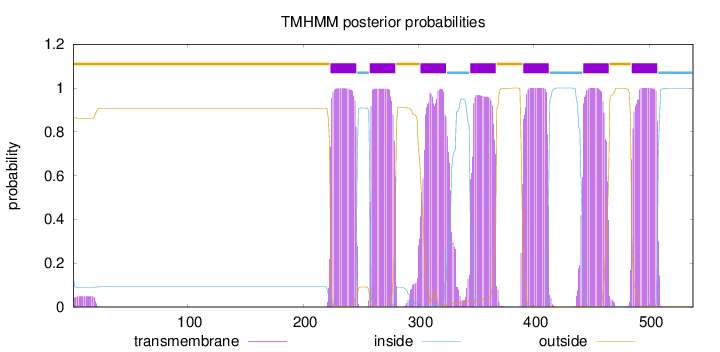
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
160.58461
Exp number, first 60 AAs:
0.910589999999999
Total prob of N-in:
0.13468
outside
1 - 223
TMhelix
224 - 246
inside
247 - 257
TMhelix
258 - 280
outside
281 - 301
TMhelix
302 - 324
inside
325 - 344
TMhelix
345 - 367
outside
368 - 390
TMhelix
391 - 413
inside
414 - 442
TMhelix
443 - 465
outside
466 - 484
TMhelix
485 - 507
inside
508 - 538
Population Genetic Test Statistics
Pi
21.194689
Theta
20.448443
Tajima's D
0.137528
CLR
0.753708
CSRT
0.414179291035448
Interpretation
Uncertain
