Gene
KWMTBOMO06917 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014474
Annotation
PREDICTED:_cytosolic_Fe-S_cluster_assembly_factor_NUBP1_homolog_[Papilio_machaon]
Full name
Cytosolic Fe-S cluster assembly factor NUBP1 homolog
+ More
Cytosolic Fe-S cluster assembly factor NUBP1
Cytosolic Fe-S cluster assembly factor nubp1
Cytosolic Fe-S cluster assembly factor NUBP1
Cytosolic Fe-S cluster assembly factor nubp1
Alternative Name
Nucleotide-binding protein 1
Location in the cell
Cytoplasmic Reliability : 1.361 Extracellular Reliability : 1.034
Sequence
CDS
ATGAGTCCTGATATAAACGTAGGTATACTCGATGCCGACATCTGCGGCCCGAGCCAGCCGCGAGTGTTGGGCGTCCGCGGAGAGCAGGTCCACAACTCCGGCTCGGGGTGGTCCCCTGTTTACGTTACGGATAACTTGTCGCTGATGTCGATCGGGTTCCTGTTGGGCAGTCCCGACGACGCCGTCATATGGAGGGGACCGAAGAAGAACGGTATGATCAAGCAGTTCCTGAGCGAAGTGGACTGGGGAGACTTGGACTACTTGCTCATAGATACGCCTCCCGGTACGTCGGACGAACACCTGTCCGCGGTGCAGTACCTGGCGCAGGCGCAGGTGTCGGGCGCGCTGCTGGTCACCACGCCGCAGGAACTCTCGCTGCTCGACGTTCGCAAAGAGATTGACTTCTGTCGCAAAGTCGGGCTCAGGGTGCTCGGGGTCGTCGAGAACATGTCGCTGTTCGTGTGCCCCAACTGCAAGAGTCGCAGCGAGATCTTCCCGGCGACGTCTGGCGGGGCGGCCGCCATGTGCCGCGAGCTGCGCGTGCCGCTGCTGGGCGCGCTGCCGCTCGAGCCGCGACTGGCGCGCGCCTGCGACTCCGGCGCGCTGCCGCTGTCCGCGCCCGCCGCCGCCGCGCCCGCCGCCAACACGCACGACGCGCTGCGAGAGATCCTCGACAAAATCGTAGCAGCCTGTGAGAACTCGAACTAA
Protein
MSPDINVGILDADICGPSQPRVLGVRGEQVHNSGSGWSPVYVTDNLSLMSIGFLLGSPDDAVIWRGPKKNGMIKQFLSEVDWGDLDYLLIDTPPGTSDEHLSAVQYLAQAQVSGALLVTTPQELSLLDVRKEIDFCRKVGLRVLGVVENMSLFVCPNCKSRSEIFPATSGGAAAMCRELRVPLLGALPLEPRLARACDSGALPLSAPAAAAPAANTHDALREILDKIVAACENSN
Summary
Description
Component of the cytosolic iron-sulfur (Fe/S) protein assembly (CIA) machinery. Required for maturation of extramitochondrial Fe-S proteins. The NUBP1-NUBP2 heterotetramer forms a Fe-S scaffold complex, mediating the de novo assembly of an Fe-S cluster and its transfer to target apoproteins.
Component of the cytosolic iron-sulfur (Fe/S) protein assembly (CIA) machinery. Required for maturation of extramitochondrial Fe-S proteins. The NUBP1-NUBP2 heterotetramer forms a Fe-S scaffold complex, mediating the de novo assembly of an Fe-S cluster and its transfer to target apoproteins. Implicated in the regulation of centrosome duplication.
Component of the cytosolic iron-sulfur (Fe/S) protein assembly (CIA) machinery. Required for maturation of extramitochondrial Fe-S proteins. The NUBP1-NUBP2 heterotetramer forms a Fe-S scaffold complex, mediating the de novo assembly of an Fe-S cluster and its transfer to target apoproteins (By similarity). Implicated in the regulation of centrosome duplication (PubMed:16638812, PubMed:23807208). Negatively regulates cilium formation and structure (PubMed:23807208).
Component of the cytosolic iron-sulfur (Fe/S) protein assembly (CIA) machinery. Required for maturation of extramitochondrial Fe-S proteins. The NUBP1-NUBP2 heterotetramer forms a Fe-S scaffold complex, mediating the de novo assembly of an Fe-S cluster and its transfer to target apoproteins. Implicated in the regulation of centrosome duplication.
Component of the cytosolic iron-sulfur (Fe/S) protein assembly (CIA) machinery. Required for maturation of extramitochondrial Fe-S proteins. The NUBP1-NUBP2 heterotetramer forms a Fe-S scaffold complex, mediating the de novo assembly of an Fe-S cluster and its transfer to target apoproteins (By similarity). Implicated in the regulation of centrosome duplication (PubMed:16638812, PubMed:23807208). Negatively regulates cilium formation and structure (PubMed:23807208).
Cofactor
[4Fe-4S] cluster
Subunit
Heterotetramer of 2 NUBP1 and 2 NUBP2 chains.
Heterotetramer of 2 NUBP1 and 2 NUBP2 chains. Interacts with KIFC1.
Heterotetramer of 2 NUBP1 and 2 NUBP2 chains (PubMed:16638812). Interacts with KIFC1 (PubMed:16638812). Interacts with NUBP2 (PubMed:16638812). Interacts with the BBS/CCT complex subunit CCT1 (PubMed:23807208).
Heterotetramer of 2 NUBP1 and 2 NUBP2 chains. Interacts with KIFC1.
Heterotetramer of 2 NUBP1 and 2 NUBP2 chains (PubMed:16638812). Interacts with KIFC1 (PubMed:16638812). Interacts with NUBP2 (PubMed:16638812). Interacts with the BBS/CCT complex subunit CCT1 (PubMed:23807208).
Miscellaneous
NIH3T3 cells expressing reduced levels of Nubp1 show an increase in number of centrosomes per cell and an increase in the fraction of multinucleated cells.
Similarity
Belongs to the Mrp/NBP35 ATP-binding proteins family. NUBP1/NBP35 subfamily.
Keywords
4Fe-4S
ATP-binding
Complete proteome
Cytoplasm
Iron
Iron-sulfur
Metal-binding
Nucleotide-binding
Reference proteome
Acetylation
Cell projection
Cilium
Cytoskeleton
Nucleus
Phosphoprotein
Feature
chain Cytosolic Fe-S cluster assembly factor NUBP1 homolog
Uniprot
H9JY56
A0A194QV59
S4PDY4
A0A2A4K6W7
A0A2H1V348
A0A194QB06
+ More
A0A2W1BXA9 A0A212FLU1 A0A1Y1N8H1 D6WEX4 E2BJD8 A0A3S2LPW6 A0A2J7RA13 E2A591 A0A088AP39 E9INW3 F4WH37 A0A0C9S349 A0A1S3HLM9 A0A182HD54 A0A151WZL2 A0A0J7L320 V9IEJ7 A0A067R403 A0A232EPM2 K7IPK4 A0A2P8XCN6 Q16T79 A0A3Q3E281 A0A091CRV9 I3KF97 G5BWF6 A0A2R5LIE4 A0A3B5B3B2 A0A3Q1EHX0 G3PMZ2 A0A084W4E7 D1MLM1 A0A182MGA4 A0A3B3Z9N6 H0W187 A0A2I4B3T3 A0A182RMQ8 U5EYU8 W5MGB5 A0A1S4FQ10 A0A3B4Y2K8 A0A3Q1B7W4 A0A182W259 A0A3Q3GCI6 A0A3Q1G533 A0A1W4WPT3 A0A3B3DX84 A0A3P8T4Q6 A0A3Q1HDB2 I3MY91 A0A3Q3R552 R7VHY1 A0A0S3NTE7 A0A3B4USC2 A0A182PK96 A0A3Q7Y588 A0A3P8T2C5 A0A1A7XPG7 A0A3Q7MW19 A0A1A8L738 A0A2K6GJ31 A0A182K8S5 D2HK11 A0A182J080 A0A1A8E696 A0A1A8UZ86 A0A1A8BTJ5 A0A1A8K4C0 A0A3Q3RNR9 A0A1B6IMI1 A0A3L7HBZ0 I3LTH4 G3GWK1 G1SEC3 H2TRT0 A0A1A8N5N0 A0A2D4NHN9 Q9R060 A0A3Q0SHA6 A7RUD5 A0A1Z5L5U2 A0A1U7TDL7 A0A1A8HB21 A0A2Y9HV17 A0A3P9Q214 A0A1Q3FHY3 H2N1L2 G1M8I1 A0A182KLL8 A0A3Q3B5R7 A0A336MB61 A0A336MY61 A0A3B3YE77 A0A336MB13
A0A2W1BXA9 A0A212FLU1 A0A1Y1N8H1 D6WEX4 E2BJD8 A0A3S2LPW6 A0A2J7RA13 E2A591 A0A088AP39 E9INW3 F4WH37 A0A0C9S349 A0A1S3HLM9 A0A182HD54 A0A151WZL2 A0A0J7L320 V9IEJ7 A0A067R403 A0A232EPM2 K7IPK4 A0A2P8XCN6 Q16T79 A0A3Q3E281 A0A091CRV9 I3KF97 G5BWF6 A0A2R5LIE4 A0A3B5B3B2 A0A3Q1EHX0 G3PMZ2 A0A084W4E7 D1MLM1 A0A182MGA4 A0A3B3Z9N6 H0W187 A0A2I4B3T3 A0A182RMQ8 U5EYU8 W5MGB5 A0A1S4FQ10 A0A3B4Y2K8 A0A3Q1B7W4 A0A182W259 A0A3Q3GCI6 A0A3Q1G533 A0A1W4WPT3 A0A3B3DX84 A0A3P8T4Q6 A0A3Q1HDB2 I3MY91 A0A3Q3R552 R7VHY1 A0A0S3NTE7 A0A3B4USC2 A0A182PK96 A0A3Q7Y588 A0A3P8T2C5 A0A1A7XPG7 A0A3Q7MW19 A0A1A8L738 A0A2K6GJ31 A0A182K8S5 D2HK11 A0A182J080 A0A1A8E696 A0A1A8UZ86 A0A1A8BTJ5 A0A1A8K4C0 A0A3Q3RNR9 A0A1B6IMI1 A0A3L7HBZ0 I3LTH4 G3GWK1 G1SEC3 H2TRT0 A0A1A8N5N0 A0A2D4NHN9 Q9R060 A0A3Q0SHA6 A7RUD5 A0A1Z5L5U2 A0A1U7TDL7 A0A1A8HB21 A0A2Y9HV17 A0A3P9Q214 A0A1Q3FHY3 H2N1L2 G1M8I1 A0A182KLL8 A0A3Q3B5R7 A0A336MB61 A0A336MY61 A0A3B3YE77 A0A336MB13
Pubmed
19121390
26354079
23622113
28756777
22118469
28004739
+ More
18362917 19820115 20798317 21282665 21719571 26483478 24845553 28648823 20075255 29403074 17510324 25186727 21993625 24438588 20920202 25463417 21993624 29451363 23254933 26514418 20010809 29704459 21804562 21551351 10486206 16141072 15489334 16638812 21183079 23807208 17615350 28528879 17554307 20966253
18362917 19820115 20798317 21282665 21719571 26483478 24845553 28648823 20075255 29403074 17510324 25186727 21993625 24438588 20920202 25463417 21993624 29451363 23254933 26514418 20010809 29704459 21804562 21551351 10486206 16141072 15489334 16638812 21183079 23807208 17615350 28528879 17554307 20966253
EMBL
BABH01042256
BABH01042257
KQ461185
KPJ07441.1
GAIX01003441
JAA89119.1
+ More
NWSH01000092 PCG79654.1 ODYU01000452 SOQ35270.1 KQ459252 KPJ02175.1 KZ149906 PZC78285.1 AGBW02007696 OWR54713.1 GEZM01013421 JAV92576.1 KQ971318 EFA00451.1 GL448558 EFN84288.1 RSAL01000034 RVE51383.1 NEVH01006580 PNF37673.1 GL436894 EFN71396.1 GL764397 EFZ17748.1 GL888148 EGI66516.1 GBYB01015341 JAG85108.1 JXUM01033983 KQ561007 KXJ80063.1 KQ982636 KYQ53309.1 LBMM01001148 KMQ96824.1 JR040269 AEY59057.1 KK852999 KDR12672.1 NNAY01002918 OXU20310.1 PYGN01002997 PSN29762.1 CH477658 KN124499 KFO20982.1 AERX01035694 AERX01035695 JH172216 GEBF01005148 EHB13617.1 JAN98484.1 GGLE01005174 MBY09300.1 ATLV01020328 KE525298 KFB45091.1 GU196316 ACZ26272.1 AXCM01001644 AAKN02007515 GANO01001800 JAB58071.1 AHAT01013584 AGTP01047032 AMQN01004585 KB293867 ELU15310.1 AB984715 BAT46579.1 HADW01018592 HADX01003809 SBP19992.1 HAEF01003253 HAEG01000497 SBR40635.1 GL192934 EFB25174.1 HAEA01012581 SBQ41061.1 HADY01010735 HAEJ01012190 SBS52647.1 HADZ01005904 SBP69845.1 HAED01022201 HAEE01007148 SBR27168.1 GECU01034955 GECU01030666 GECU01027094 GECU01026824 GECU01024582 GECU01021566 GECU01019587 GECU01007020 GECU01002594 GECU01002361 JAS72751.1 JAS77040.1 JAS80612.1 JAS80882.1 JAS83124.1 JAS86140.1 JAS88119.1 JAT00687.1 JAT05113.1 JAT05346.1 RAZU01000238 RLQ63458.1 AEMK02000018 JH000052 EGV96311.1 AAGW02056958 AAGW02056959 HAEH01000191 HAEI01006838 SBR64390.1 IACM01181825 LAB45230.1 AF114170 AK020141 AK157212 BC055436 DS469540 GFJQ02004184 JAW02786.1 HAEB01002035 HAEC01012197 SBQ80414.1 GFDL01007864 JAV27181.1 ACTA01137194 ACTA01145193 UFQT01000836 SSX27525.1 UFQT01001491 SSX30798.1 SSX27524.1
NWSH01000092 PCG79654.1 ODYU01000452 SOQ35270.1 KQ459252 KPJ02175.1 KZ149906 PZC78285.1 AGBW02007696 OWR54713.1 GEZM01013421 JAV92576.1 KQ971318 EFA00451.1 GL448558 EFN84288.1 RSAL01000034 RVE51383.1 NEVH01006580 PNF37673.1 GL436894 EFN71396.1 GL764397 EFZ17748.1 GL888148 EGI66516.1 GBYB01015341 JAG85108.1 JXUM01033983 KQ561007 KXJ80063.1 KQ982636 KYQ53309.1 LBMM01001148 KMQ96824.1 JR040269 AEY59057.1 KK852999 KDR12672.1 NNAY01002918 OXU20310.1 PYGN01002997 PSN29762.1 CH477658 KN124499 KFO20982.1 AERX01035694 AERX01035695 JH172216 GEBF01005148 EHB13617.1 JAN98484.1 GGLE01005174 MBY09300.1 ATLV01020328 KE525298 KFB45091.1 GU196316 ACZ26272.1 AXCM01001644 AAKN02007515 GANO01001800 JAB58071.1 AHAT01013584 AGTP01047032 AMQN01004585 KB293867 ELU15310.1 AB984715 BAT46579.1 HADW01018592 HADX01003809 SBP19992.1 HAEF01003253 HAEG01000497 SBR40635.1 GL192934 EFB25174.1 HAEA01012581 SBQ41061.1 HADY01010735 HAEJ01012190 SBS52647.1 HADZ01005904 SBP69845.1 HAED01022201 HAEE01007148 SBR27168.1 GECU01034955 GECU01030666 GECU01027094 GECU01026824 GECU01024582 GECU01021566 GECU01019587 GECU01007020 GECU01002594 GECU01002361 JAS72751.1 JAS77040.1 JAS80612.1 JAS80882.1 JAS83124.1 JAS86140.1 JAS88119.1 JAT00687.1 JAT05113.1 JAT05346.1 RAZU01000238 RLQ63458.1 AEMK02000018 JH000052 EGV96311.1 AAGW02056958 AAGW02056959 HAEH01000191 HAEI01006838 SBR64390.1 IACM01181825 LAB45230.1 AF114170 AK020141 AK157212 BC055436 DS469540 GFJQ02004184 JAW02786.1 HAEB01002035 HAEC01012197 SBQ80414.1 GFDL01007864 JAV27181.1 ACTA01137194 ACTA01145193 UFQT01000836 SSX27525.1 UFQT01001491 SSX30798.1 SSX27524.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000007266
+ More
UP000008237 UP000283053 UP000235965 UP000000311 UP000005203 UP000007755 UP000085678 UP000069940 UP000249989 UP000075809 UP000036403 UP000027135 UP000215335 UP000002358 UP000245037 UP000008820 UP000264820 UP000028990 UP000005207 UP000006813 UP000261400 UP000257200 UP000007635 UP000030765 UP000075883 UP000261520 UP000005447 UP000192220 UP000075900 UP000018468 UP000261360 UP000257160 UP000075920 UP000261660 UP000192223 UP000261560 UP000265080 UP000265040 UP000005215 UP000261600 UP000014760 UP000261420 UP000075885 UP000286642 UP000286641 UP000233160 UP000075881 UP000075880 UP000261640 UP000273346 UP000008227 UP000001075 UP000001811 UP000005226 UP000000589 UP000261340 UP000001593 UP000189704 UP000248481 UP000242638 UP000001038 UP000008912 UP000075882 UP000264800 UP000261480
UP000008237 UP000283053 UP000235965 UP000000311 UP000005203 UP000007755 UP000085678 UP000069940 UP000249989 UP000075809 UP000036403 UP000027135 UP000215335 UP000002358 UP000245037 UP000008820 UP000264820 UP000028990 UP000005207 UP000006813 UP000261400 UP000257200 UP000007635 UP000030765 UP000075883 UP000261520 UP000005447 UP000192220 UP000075900 UP000018468 UP000261360 UP000257160 UP000075920 UP000261660 UP000192223 UP000261560 UP000265080 UP000265040 UP000005215 UP000261600 UP000014760 UP000261420 UP000075885 UP000286642 UP000286641 UP000233160 UP000075881 UP000075880 UP000261640 UP000273346 UP000008227 UP000001075 UP000001811 UP000005226 UP000000589 UP000261340 UP000001593 UP000189704 UP000248481 UP000242638 UP000001038 UP000008912 UP000075882 UP000264800 UP000261480
Pfam
PF10609 ParA
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9JY56
A0A194QV59
S4PDY4
A0A2A4K6W7
A0A2H1V348
A0A194QB06
+ More
A0A2W1BXA9 A0A212FLU1 A0A1Y1N8H1 D6WEX4 E2BJD8 A0A3S2LPW6 A0A2J7RA13 E2A591 A0A088AP39 E9INW3 F4WH37 A0A0C9S349 A0A1S3HLM9 A0A182HD54 A0A151WZL2 A0A0J7L320 V9IEJ7 A0A067R403 A0A232EPM2 K7IPK4 A0A2P8XCN6 Q16T79 A0A3Q3E281 A0A091CRV9 I3KF97 G5BWF6 A0A2R5LIE4 A0A3B5B3B2 A0A3Q1EHX0 G3PMZ2 A0A084W4E7 D1MLM1 A0A182MGA4 A0A3B3Z9N6 H0W187 A0A2I4B3T3 A0A182RMQ8 U5EYU8 W5MGB5 A0A1S4FQ10 A0A3B4Y2K8 A0A3Q1B7W4 A0A182W259 A0A3Q3GCI6 A0A3Q1G533 A0A1W4WPT3 A0A3B3DX84 A0A3P8T4Q6 A0A3Q1HDB2 I3MY91 A0A3Q3R552 R7VHY1 A0A0S3NTE7 A0A3B4USC2 A0A182PK96 A0A3Q7Y588 A0A3P8T2C5 A0A1A7XPG7 A0A3Q7MW19 A0A1A8L738 A0A2K6GJ31 A0A182K8S5 D2HK11 A0A182J080 A0A1A8E696 A0A1A8UZ86 A0A1A8BTJ5 A0A1A8K4C0 A0A3Q3RNR9 A0A1B6IMI1 A0A3L7HBZ0 I3LTH4 G3GWK1 G1SEC3 H2TRT0 A0A1A8N5N0 A0A2D4NHN9 Q9R060 A0A3Q0SHA6 A7RUD5 A0A1Z5L5U2 A0A1U7TDL7 A0A1A8HB21 A0A2Y9HV17 A0A3P9Q214 A0A1Q3FHY3 H2N1L2 G1M8I1 A0A182KLL8 A0A3Q3B5R7 A0A336MB61 A0A336MY61 A0A3B3YE77 A0A336MB13
A0A2W1BXA9 A0A212FLU1 A0A1Y1N8H1 D6WEX4 E2BJD8 A0A3S2LPW6 A0A2J7RA13 E2A591 A0A088AP39 E9INW3 F4WH37 A0A0C9S349 A0A1S3HLM9 A0A182HD54 A0A151WZL2 A0A0J7L320 V9IEJ7 A0A067R403 A0A232EPM2 K7IPK4 A0A2P8XCN6 Q16T79 A0A3Q3E281 A0A091CRV9 I3KF97 G5BWF6 A0A2R5LIE4 A0A3B5B3B2 A0A3Q1EHX0 G3PMZ2 A0A084W4E7 D1MLM1 A0A182MGA4 A0A3B3Z9N6 H0W187 A0A2I4B3T3 A0A182RMQ8 U5EYU8 W5MGB5 A0A1S4FQ10 A0A3B4Y2K8 A0A3Q1B7W4 A0A182W259 A0A3Q3GCI6 A0A3Q1G533 A0A1W4WPT3 A0A3B3DX84 A0A3P8T4Q6 A0A3Q1HDB2 I3MY91 A0A3Q3R552 R7VHY1 A0A0S3NTE7 A0A3B4USC2 A0A182PK96 A0A3Q7Y588 A0A3P8T2C5 A0A1A7XPG7 A0A3Q7MW19 A0A1A8L738 A0A2K6GJ31 A0A182K8S5 D2HK11 A0A182J080 A0A1A8E696 A0A1A8UZ86 A0A1A8BTJ5 A0A1A8K4C0 A0A3Q3RNR9 A0A1B6IMI1 A0A3L7HBZ0 I3LTH4 G3GWK1 G1SEC3 H2TRT0 A0A1A8N5N0 A0A2D4NHN9 Q9R060 A0A3Q0SHA6 A7RUD5 A0A1Z5L5U2 A0A1U7TDL7 A0A1A8HB21 A0A2Y9HV17 A0A3P9Q214 A0A1Q3FHY3 H2N1L2 G1M8I1 A0A182KLL8 A0A3Q3B5R7 A0A336MB61 A0A336MY61 A0A3B3YE77 A0A336MB13
PDB
6G2G
E-value=3.72048e-53,
Score=524
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Cell projection Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Cytoskeleton Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Cilium axoneme Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Cilium basal body Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Microtubule organizing center Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Centrosome Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Centriole Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Nucleus Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Cell projection Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Cytoskeleton Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Cilium axoneme Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Cilium basal body Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Microtubule organizing center Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Centrosome Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
Centriole Enriched in centrioles of microtubule asters during prophase, prometaphase and telophase stages of mitosis. Localized at centrioles and in the nucleus at interphase. Colocalizes with NUBP2 at prometaphase. Specifically localizes to the axenome of motile cilia as opposed to primary non-motile cilia. Localization is independent of NUBP2 and KIFC1. With evidence from 5 publications.
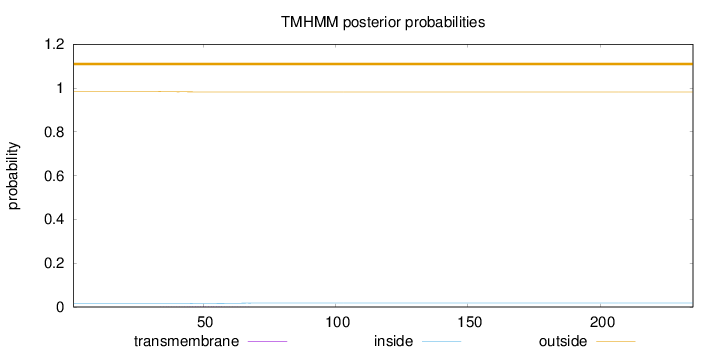
Length:
235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0415
Exp number, first 60 AAs:
0.03451
Total prob of N-in:
0.01650
outside
1 - 235
Population Genetic Test Statistics
Pi
18.450131
Theta
17.169232
Tajima's D
0.570863
CLR
1.435295
CSRT
0.547072646367682
Interpretation
Uncertain
