Gene
KWMTBOMO06916
Pre Gene Modal
BGIBMGA014475
Annotation
hypothetical_protein_KGM_10330_[Danaus_plexippus]
Full name
Sulfhydryl oxidase
+ More
Multifunctional fusion protein
Multifunctional fusion protein
Location in the cell
Nuclear Reliability : 2.765
Sequence
CDS
ATGCCTCCACACCACGAGGAAGAAGAAGATAAACCTTGTAGAGCATGTTCGGATTTCAAAACTTGGGCAAAACTACAAAACAAAGGTTCAAAAACCGCGCCGACCAAACCGCCTCCTCCTCAATCAAAGGAATGCCCTCTAGATAAAGAAGAATTAGGTAAATCTACATGGGGTTTCCTCCACTCGATGGCTTCATATTTTCCTGACAAGCCAACTAAATTACAATCGGAAAATATGTCACGATTCTTTAATATTTTTGCCCAATTCTATCCATGTGAACCATGTTCTTTGGACTTTCTGGATGACATACAAAAGAACCCACCAAAGACAAAATCAAGGGATGAACTAGCAAAATGGCTCTGTGAGAGACACAATACTGTGAATGTAAAACTTGGAAAACCATTATTTGACTGTAGCAAAATTCACGAGAGGTGGAGAGATGGGTGGAAAGATGGCTCCTGTGATTAA
Protein
MPPHHEEEEDKPCRACSDFKTWAKLQNKGSKTAPTKPPPPQSKECPLDKEELGKSTWGFLHSMASYFPDKPTKLQSENMSRFFNIFAQFYPCEPCSLDFLDDIQKNPPKTKSRDELAKWLCERHNTVNVKLGKPLFDCSKIHERWRDGWKDGSCD
Summary
Catalytic Activity
O2 + 2 R'C(R)SH = H2O2 + R'C(R)S-S(R)CR'
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
Cofactor
FAD
Similarity
Belongs to the PPP phosphatase family.
Uniprot
S4PFG6
A0A2W1BXD2
A0A2A4K7A9
A0A2H1V395
A0A212FLU7
A0A1E1WDE6
+ More
A0A194QPL0 A0A194Q9T1 H3CAD8 Q4TBL6 A0A3B4D272 M3ZGW2 A0A3Q4GFZ4 A0A3Q2WS86 A0A3P8PMV3 A0A3P9BD80 A0A3B4GSY1 A0A3Q3VQJ6 A0A3P9Q5F3 A0A1A7YYZ3 A0A2D0PZJ9 A0A0S7ICH5 A0A3Q3VWQ0 A0A3B3UKB1 A0A3P8WPZ4 H2RVL7 A0A3Q2XBK2 A0A3B5ATA1 A0A3B5PUF9 A0A3Q3B2N5 A0A2U9CSL6 A0A3B3U7J5 G3PMJ9 A0A3Q2U2E1 E0VYU4 A0A3Q3G9T9 A0A3Q1BR50 A0A232EWS7 A0A3P8RRM4 A0A3Q3GAE8 A0A146VZ23 W5KSE0 A0A3Q2G7A8 A0A3Q3KZS4 K7JHH1 A0A146SA48 A0A3B4Y7V3 A0A3B4T845 A0A1A8FVA9 A0A1A8I0E6 A0A151N2H6 A0A2G8JPK6 A0A3Q1EQA7 A0A1A8Q6V9 A0A3B3CQ66 I3KF07 A0A091QWY3 A0A1A8P7X1 A0A1A8E932 A0A146ZXG9 A0A1S3QD91 A0A1A8BDX4 H0Z856 A4QNU9 A0A1A8AUT8 A0A3Q3R2V4 A0A0F8BMU5 A0A087XPE9 A0A0F8ATH6 A0A3B3TBI6 C3Z7T7 V5H6F6 A0A3B4AP22 C1BJH2 A0A1L8EYL9 H3AS15 W5MG16 A0A3Q1IVL0 U5ERT6 A0A3B3WHC2 A0A087XNT9 A0A0A9VZL1 A0A2I4B3N9 A0A2R7WJE4 A0A0L8I3Y5 G3MJ32 A0A2L2YMN3 Q4V7G7 A0A1S3QLR1 A0A226NMS8 Q16FJ5 A0A182NJ22 A0A0J7KQF5 H2MCW5 A0A1J1J3N9 A0A1S3J937 A0A026WQM8 A0A226PQV8 A0A218U9R6
A0A194QPL0 A0A194Q9T1 H3CAD8 Q4TBL6 A0A3B4D272 M3ZGW2 A0A3Q4GFZ4 A0A3Q2WS86 A0A3P8PMV3 A0A3P9BD80 A0A3B4GSY1 A0A3Q3VQJ6 A0A3P9Q5F3 A0A1A7YYZ3 A0A2D0PZJ9 A0A0S7ICH5 A0A3Q3VWQ0 A0A3B3UKB1 A0A3P8WPZ4 H2RVL7 A0A3Q2XBK2 A0A3B5ATA1 A0A3B5PUF9 A0A3Q3B2N5 A0A2U9CSL6 A0A3B3U7J5 G3PMJ9 A0A3Q2U2E1 E0VYU4 A0A3Q3G9T9 A0A3Q1BR50 A0A232EWS7 A0A3P8RRM4 A0A3Q3GAE8 A0A146VZ23 W5KSE0 A0A3Q2G7A8 A0A3Q3KZS4 K7JHH1 A0A146SA48 A0A3B4Y7V3 A0A3B4T845 A0A1A8FVA9 A0A1A8I0E6 A0A151N2H6 A0A2G8JPK6 A0A3Q1EQA7 A0A1A8Q6V9 A0A3B3CQ66 I3KF07 A0A091QWY3 A0A1A8P7X1 A0A1A8E932 A0A146ZXG9 A0A1S3QD91 A0A1A8BDX4 H0Z856 A4QNU9 A0A1A8AUT8 A0A3Q3R2V4 A0A0F8BMU5 A0A087XPE9 A0A0F8ATH6 A0A3B3TBI6 C3Z7T7 V5H6F6 A0A3B4AP22 C1BJH2 A0A1L8EYL9 H3AS15 W5MG16 A0A3Q1IVL0 U5ERT6 A0A3B3WHC2 A0A087XNT9 A0A0A9VZL1 A0A2I4B3N9 A0A2R7WJE4 A0A0L8I3Y5 G3MJ32 A0A2L2YMN3 Q4V7G7 A0A1S3QLR1 A0A226NMS8 Q16FJ5 A0A182NJ22 A0A0J7KQF5 H2MCW5 A0A1J1J3N9 A0A1S3J937 A0A026WQM8 A0A226PQV8 A0A218U9R6
EC Number
1.8.3.2
Pubmed
23622113
28756777
22118469
26354079
15496914
23542700
+ More
25186727 24487278 21551351 20566863 28648823 25329095 20075255 22293439 29023486 29451363 20360741 23594743 25835551 29240929 18563158 25765539 25463417 27762356 9215903 25401762 26823975 22216098 26561354 17510324 17554307 24508170 30249741 24621616
25186727 24487278 21551351 20566863 28648823 25329095 20075255 22293439 29023486 29451363 20360741 23594743 25835551 29240929 18563158 25765539 25463417 27762356 9215903 25401762 26823975 22216098 26561354 17510324 17554307 24508170 30249741 24621616
EMBL
GAIX01006600
JAA85960.1
KZ149906
PZC78284.1
NWSH01000092
PCG79653.1
+ More
ODYU01000452 SOQ35269.1 AGBW02007696 OWR54714.1 GDQN01006097 JAT84957.1 KQ461185 KPJ07442.1 KQ459252 KPJ02174.1 CAAE01007118 CAF89716.1 HADW01006811 HADX01012918 SBP35150.1 GBYX01430494 JAO50806.1 CP026261 AWP18666.1 DS235848 EEB18550.1 NNAY01001823 OXU22804.1 GCES01064086 JAR22237.1 GCES01108888 JAQ77434.1 HAEB01016400 HAEC01004024 SBQ62927.1 HAED01005109 HAEE01014043 SBQ91139.1 AKHW03004153 KYO30845.1 MRZV01001473 PIK37628.1 HAEF01004276 HAEG01011154 SBR88987.1 AERX01035671 KK805407 KFQ31723.1 HAEI01001657 HAEH01017301 SBR77112.1 HAEA01013129 SBQ41609.1 GCES01015658 JAR70665.1 HADZ01001826 SBP65767.1 ABQF01123828 BX649547 BC139530 AAI39531.1 HADY01019520 HAEJ01012724 SBP58005.1 SBS53181.1 KQ042629 KKF12958.1 AYCK01010978 AYCK01010979 AYCK01010980 KQ041130 KKF30230.1 GG666591 EEN51442.1 GANP01005858 JAB78610.1 BT074751 ACO09175.1 CM004482 OCT64454.1 AFYH01174689 AHAT01013578 GANO01003577 JAB56294.1 GBHO01042002 GBHO01041999 GBRD01005815 GBRD01005803 GBRD01001005 GDHC01000764 JAG01602.1 JAG01605.1 JAG60006.1 JAQ17865.1 KK854912 PTY19629.1 KQ416641 KOF96069.1 JO841883 AEO33500.1 IAAA01034264 LAA09398.1 BC097922 AAH97922.1 MCFN01000008 OXB68801.1 CH478424 CH477539 EAT33007.1 EAT39297.1 LBMM01004318 KMQ92572.1 CVRI01000069 CRL06999.1 KK107128 QOIP01000006 EZA58330.1 RLU21649.1 AWGT02000031 OXB81882.1 MUZQ01000541 OWK50479.1
ODYU01000452 SOQ35269.1 AGBW02007696 OWR54714.1 GDQN01006097 JAT84957.1 KQ461185 KPJ07442.1 KQ459252 KPJ02174.1 CAAE01007118 CAF89716.1 HADW01006811 HADX01012918 SBP35150.1 GBYX01430494 JAO50806.1 CP026261 AWP18666.1 DS235848 EEB18550.1 NNAY01001823 OXU22804.1 GCES01064086 JAR22237.1 GCES01108888 JAQ77434.1 HAEB01016400 HAEC01004024 SBQ62927.1 HAED01005109 HAEE01014043 SBQ91139.1 AKHW03004153 KYO30845.1 MRZV01001473 PIK37628.1 HAEF01004276 HAEG01011154 SBR88987.1 AERX01035671 KK805407 KFQ31723.1 HAEI01001657 HAEH01017301 SBR77112.1 HAEA01013129 SBQ41609.1 GCES01015658 JAR70665.1 HADZ01001826 SBP65767.1 ABQF01123828 BX649547 BC139530 AAI39531.1 HADY01019520 HAEJ01012724 SBP58005.1 SBS53181.1 KQ042629 KKF12958.1 AYCK01010978 AYCK01010979 AYCK01010980 KQ041130 KKF30230.1 GG666591 EEN51442.1 GANP01005858 JAB78610.1 BT074751 ACO09175.1 CM004482 OCT64454.1 AFYH01174689 AHAT01013578 GANO01003577 JAB56294.1 GBHO01042002 GBHO01041999 GBRD01005815 GBRD01005803 GBRD01001005 GDHC01000764 JAG01602.1 JAG01605.1 JAG60006.1 JAQ17865.1 KK854912 PTY19629.1 KQ416641 KOF96069.1 JO841883 AEO33500.1 IAAA01034264 LAA09398.1 BC097922 AAH97922.1 MCFN01000008 OXB68801.1 CH478424 CH477539 EAT33007.1 EAT39297.1 LBMM01004318 KMQ92572.1 CVRI01000069 CRL06999.1 KK107128 QOIP01000006 EZA58330.1 RLU21649.1 AWGT02000031 OXB81882.1 MUZQ01000541 OWK50479.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000007303
UP000261440
+ More
UP000002852 UP000261580 UP000264840 UP000265100 UP000265160 UP000261460 UP000261620 UP000242638 UP000221080 UP000261500 UP000265120 UP000005226 UP000264820 UP000261400 UP000264800 UP000246464 UP000007635 UP000265000 UP000009046 UP000261660 UP000257160 UP000215335 UP000265080 UP000018467 UP000265020 UP000261640 UP000002358 UP000261360 UP000261420 UP000050525 UP000230750 UP000257200 UP000261560 UP000005207 UP000087266 UP000007754 UP000000437 UP000261600 UP000028760 UP000261540 UP000001554 UP000261520 UP000186698 UP000008672 UP000018468 UP000265040 UP000261480 UP000192220 UP000053454 UP000198323 UP000008820 UP000075884 UP000036403 UP000001038 UP000183832 UP000085678 UP000053097 UP000279307 UP000198419 UP000197619
UP000002852 UP000261580 UP000264840 UP000265100 UP000265160 UP000261460 UP000261620 UP000242638 UP000221080 UP000261500 UP000265120 UP000005226 UP000264820 UP000261400 UP000264800 UP000246464 UP000007635 UP000265000 UP000009046 UP000261660 UP000257160 UP000215335 UP000265080 UP000018467 UP000265020 UP000261640 UP000002358 UP000261360 UP000261420 UP000050525 UP000230750 UP000257200 UP000261560 UP000005207 UP000087266 UP000007754 UP000000437 UP000261600 UP000028760 UP000261540 UP000001554 UP000261520 UP000186698 UP000008672 UP000018468 UP000265040 UP000261480 UP000192220 UP000053454 UP000198323 UP000008820 UP000075884 UP000036403 UP000001038 UP000183832 UP000085678 UP000053097 UP000279307 UP000198419 UP000197619
Interpro
SUPFAM
SSF69000
SSF69000
Gene 3D
ProteinModelPortal
S4PFG6
A0A2W1BXD2
A0A2A4K7A9
A0A2H1V395
A0A212FLU7
A0A1E1WDE6
+ More
A0A194QPL0 A0A194Q9T1 H3CAD8 Q4TBL6 A0A3B4D272 M3ZGW2 A0A3Q4GFZ4 A0A3Q2WS86 A0A3P8PMV3 A0A3P9BD80 A0A3B4GSY1 A0A3Q3VQJ6 A0A3P9Q5F3 A0A1A7YYZ3 A0A2D0PZJ9 A0A0S7ICH5 A0A3Q3VWQ0 A0A3B3UKB1 A0A3P8WPZ4 H2RVL7 A0A3Q2XBK2 A0A3B5ATA1 A0A3B5PUF9 A0A3Q3B2N5 A0A2U9CSL6 A0A3B3U7J5 G3PMJ9 A0A3Q2U2E1 E0VYU4 A0A3Q3G9T9 A0A3Q1BR50 A0A232EWS7 A0A3P8RRM4 A0A3Q3GAE8 A0A146VZ23 W5KSE0 A0A3Q2G7A8 A0A3Q3KZS4 K7JHH1 A0A146SA48 A0A3B4Y7V3 A0A3B4T845 A0A1A8FVA9 A0A1A8I0E6 A0A151N2H6 A0A2G8JPK6 A0A3Q1EQA7 A0A1A8Q6V9 A0A3B3CQ66 I3KF07 A0A091QWY3 A0A1A8P7X1 A0A1A8E932 A0A146ZXG9 A0A1S3QD91 A0A1A8BDX4 H0Z856 A4QNU9 A0A1A8AUT8 A0A3Q3R2V4 A0A0F8BMU5 A0A087XPE9 A0A0F8ATH6 A0A3B3TBI6 C3Z7T7 V5H6F6 A0A3B4AP22 C1BJH2 A0A1L8EYL9 H3AS15 W5MG16 A0A3Q1IVL0 U5ERT6 A0A3B3WHC2 A0A087XNT9 A0A0A9VZL1 A0A2I4B3N9 A0A2R7WJE4 A0A0L8I3Y5 G3MJ32 A0A2L2YMN3 Q4V7G7 A0A1S3QLR1 A0A226NMS8 Q16FJ5 A0A182NJ22 A0A0J7KQF5 H2MCW5 A0A1J1J3N9 A0A1S3J937 A0A026WQM8 A0A226PQV8 A0A218U9R6
A0A194QPL0 A0A194Q9T1 H3CAD8 Q4TBL6 A0A3B4D272 M3ZGW2 A0A3Q4GFZ4 A0A3Q2WS86 A0A3P8PMV3 A0A3P9BD80 A0A3B4GSY1 A0A3Q3VQJ6 A0A3P9Q5F3 A0A1A7YYZ3 A0A2D0PZJ9 A0A0S7ICH5 A0A3Q3VWQ0 A0A3B3UKB1 A0A3P8WPZ4 H2RVL7 A0A3Q2XBK2 A0A3B5ATA1 A0A3B5PUF9 A0A3Q3B2N5 A0A2U9CSL6 A0A3B3U7J5 G3PMJ9 A0A3Q2U2E1 E0VYU4 A0A3Q3G9T9 A0A3Q1BR50 A0A232EWS7 A0A3P8RRM4 A0A3Q3GAE8 A0A146VZ23 W5KSE0 A0A3Q2G7A8 A0A3Q3KZS4 K7JHH1 A0A146SA48 A0A3B4Y7V3 A0A3B4T845 A0A1A8FVA9 A0A1A8I0E6 A0A151N2H6 A0A2G8JPK6 A0A3Q1EQA7 A0A1A8Q6V9 A0A3B3CQ66 I3KF07 A0A091QWY3 A0A1A8P7X1 A0A1A8E932 A0A146ZXG9 A0A1S3QD91 A0A1A8BDX4 H0Z856 A4QNU9 A0A1A8AUT8 A0A3Q3R2V4 A0A0F8BMU5 A0A087XPE9 A0A0F8ATH6 A0A3B3TBI6 C3Z7T7 V5H6F6 A0A3B4AP22 C1BJH2 A0A1L8EYL9 H3AS15 W5MG16 A0A3Q1IVL0 U5ERT6 A0A3B3WHC2 A0A087XNT9 A0A0A9VZL1 A0A2I4B3N9 A0A2R7WJE4 A0A0L8I3Y5 G3MJ32 A0A2L2YMN3 Q4V7G7 A0A1S3QLR1 A0A226NMS8 Q16FJ5 A0A182NJ22 A0A0J7KQF5 H2MCW5 A0A1J1J3N9 A0A1S3J937 A0A026WQM8 A0A226PQV8 A0A218U9R6
PDB
3R7C
E-value=2.06678e-34,
Score=359
Ontologies
GO
PANTHER
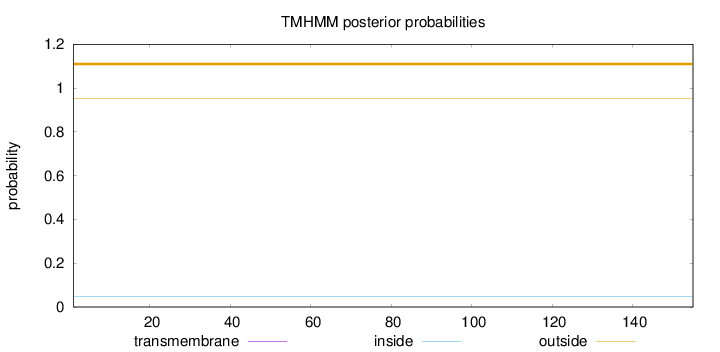
Topology
Length:
155
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0004
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04828
outside
1 - 155
Population Genetic Test Statistics
Pi
22.861561
Theta
18.272924
Tajima's D
0.76119
CLR
1.074608
CSRT
0.595620218989051
Interpretation
Uncertain
