Gene
KWMTBOMO06914
Pre Gene Modal
BGIBMGA014468
Annotation
non-LTR_retrotransposon_CATS_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 3.036
Sequence
CDS
ATGAGTAAATCGAAAGTATCAGCTGAATGGAAGAAGAGAGTTAAGTCAGAGTACATGCGGCTTCGACAAGTAAAGCGTTTCAAACGCGCTGATGAAGTGAAAGTAGCATGGGCGCGCAATTTGCAAATAATGTCCGAAGCTATAGAGATGCCTGAGGCGGAAACTACGGATCGTACTAAACGGCCCTTCTGGCCCCCGCCGGCGCCTACATCGAGCCATGAGAGCCTTATGAAGAGGGCCGAAGTTACCTATACAGACGGTTCAGGTGTAGTGACAACCCAACAGGTACCAATAAGAATAATCAACTCTGTGAATCCAATTCCGACGATGTATACATGGGCTCCAACACAGAAAAATTTTATGGTAGAAGATGAGACTGTTCTGCACAACATTCCATACATGGGAGACGAGGTCCTGGATCAGGATGGTACATTCATCGAGGAGCTAATAAAGAACTACGACGGGAAAGTGCATGGTGATAAAGAGGGGGGCTTCATAGATGACCAGCTATTTGTTGATCTGGTGCATGCATTGATGACTTACCAGACTAAAGATGATGTCAATGAAGAGAAACGAGAAAGAGAGTCAAGAAATTCTAAAGAAGATGTAGAGAAAGAAAGCAGCAAAGACAAAGATGACAAAAGCAAAGAAGATCTCCCAGAGGAAAAGAAGACAGCAGTTGAGAAACAGTTTCCGATTTTCACCATATTCCAGGCGATCAGCTTGCAGTTTCCTGACAAAGGAACAGCTCAGGAGTTAAGAGAGAATTTCGGCTTCAAGGAGCAAACCGGCACGGTCGACGCCATTAACACCCTGATAGACACTATACGAAAAGCCAAACAAGAGAGGTCACAGGTTGTCGCCGTCTCGCTCGACATCAGGGCAGCTTTTGATAACGCCTGGTGGCCGGTCCTATTACACCGCCTTAGAGATATACGATGCCCCAAAAATATCTACGGCCTCATCAAGAGCTACCTCCGAGACAGGCGCGTCGTTCTGGACCACGGAGGAGTTCGCGTCTCCAGAGCCACGTCAAAAGGCTGCGTTCAGGGCTCGGTGTGCGGGCCTGCATTGTGGAACATCATACTTGATGAACTATTGGAGATCCATCTACCATCGGGATGCCACATGCAGGCCTACGCAGACGATGTGGTACTCGTGGTCACCGCCGAAAATGTCAATCGCCTAGAGGAGCTCACCGACACGGTTCTCCAACAGATAGTGAATTGGGGAAAAAGCGTCAAGCTCGAATTCGGGGCCTCCAAAACACAACTGATCGCCTTCACCCCCAAGGCCAGAACTCTTAGGGTGGAGATGGACGGTCAAATCTTAAAAACCTGTAAGGAGATCAAGCTCCTCGGAATAATTTTGGACGAAAAGCTACTCTTTGGCCGCCATGTACAGTACGTCCTGGGAAAAGCCTCGCGAATATTCAACAAACTGTGCTTGTACTCCCGACCAACCTGGGGTGCTCATCCCGAGAACGTGCGCACCATCTATCTCCGAGTCATCGAGCCAATCGTCACCTACGCTGCCGGGGTGTGGGGACACATAGTAAACAAGCGCTACATCAAAAAATCTTTGATGAGCATGCAGCGTGGTTTTGCCCTAAAAGCCATCAGAGGATTCCGCACTATTTCAACCTCCGCTGCTCTGGCTCTCGCACAATTCACCCCTCTAGACCTAAAAATCCGAGAAGTACACCAAATCGAAAAAGTCAGGCTCACTGGGAAAACTGAATTCCTCCCCAATGACATTTCTTATGAGAAACCTGTCCCCGTCAAGGACCTCCTACATCCCGCTAGAAGGCCGACTATCAAACCCATACCCTCCTCTGAACACCGAATTAACTCCGAAATGACGGACAGTACTCTGGTCACTCGCGTATTCACTGACGGAAGTAAGCAGGACGATGGATCAGTTGGAGCCGCATTCGTGCTTTACAGACCCGGCGACACTGAAACATCCGTAACAAAAAAGTACAAATTACACAGTAGCTGCACAGTCTTTCAGGCAGAGCTGTACGCCATCTGGAAAGCCTGTCAGTGGATTGTAAGCGAAAATCATCCCCACACATACATCTTCACCGATTCTCTCTCATCAATATACGCGATAAATAACAGAAGCAACACACACCCCATCATAGTGAACATACACAAACTCATACACGCTAACCACACTACACACAAAATTTACATAGATTGGGTAAAGGGACACTCGGGTGTCATAGGAAACGAGGAAGCCGATGCGGCTGCCAACAGCGGCGCACGCATGCACAAGGCCCCCGACTACTCGCAGTTCTCAATATCCTACGTGAAACACAAGATTCGCACAGAACACCACGCCATCTGGCAAGCCAGATACGAAAGCTCCCCGCAGGGCTCACACACAAAAACACTACTACCAAAACTGAACGACATCAAAGAACTCAACAAATGCACACAAACAAACTTTCAGCTTACACAAATACTCACAGGCCATGGATATCACAAGACATACTTACATCGCTTCAAAATAGCACCAGATGACACCTGCCCCTGCGACGGCACCTCCAGCCAAACTGTAGAACACCTCCTTAAAAACTGCCAAGCGTTCGCTGCGAAAAGGCACAGTCACGAAGAAACGTGCCACGCAGTACAGGTGCCACCATATAATTTACCTGAAATGTTAAAAAAGAAAGCTGCAATCAACTCTTTCACCTCGTTTTGCCAAATAATAATCAACAACATCAAGAAAATAAATGAAAACAATTAG
Protein
MSKSKVSAEWKKRVKSEYMRLRQVKRFKRADEVKVAWARNLQIMSEAIEMPEAETTDRTKRPFWPPPAPTSSHESLMKRAEVTYTDGSGVVTTQQVPIRIINSVNPIPTMYTWAPTQKNFMVEDETVLHNIPYMGDEVLDQDGTFIEELIKNYDGKVHGDKEGGFIDDQLFVDLVHALMTYQTKDDVNEEKRERESRNSKEDVEKESSKDKDDKSKEDLPEEKKTAVEKQFPIFTIFQAISLQFPDKGTAQELRENFGFKEQTGTVDAINTLIDTIRKAKQERSQVVAVSLDIRAAFDNAWWPVLLHRLRDIRCPKNIYGLIKSYLRDRRVVLDHGGVRVSRATSKGCVQGSVCGPALWNIILDELLEIHLPSGCHMQAYADDVVLVVTAENVNRLEELTDTVLQQIVNWGKSVKLEFGASKTQLIAFTPKARTLRVEMDGQILKTCKEIKLLGIILDEKLLFGRHVQYVLGKASRIFNKLCLYSRPTWGAHPENVRTIYLRVIEPIVTYAAGVWGHIVNKRYIKKSLMSMQRGFALKAIRGFRTISTSAALALAQFTPLDLKIREVHQIEKVRLTGKTEFLPNDISYEKPVPVKDLLHPARRPTIKPIPSSEHRINSEMTDSTLVTRVFTDGSKQDDGSVGAAFVLYRPGDTETSVTKKYKLHSSCTVFQAELYAIWKACQWIVSENHPHTYIFTDSLSSIYAINNRSNTHPIIVNIHKLIHANHTTHKIYIDWVKGHSGVIGNEEADAAANSGARMHKAPDYSQFSISYVKHKIRTEHHAIWQARYESSPQGSHTKTLLPKLNDIKELNKCTQTNFQLTQILTGHGYHKTYLHRFKIAPDDTCPCDGTSSQTVEHLLKNCQAFAAKRHSHEETCHAVQVPPYNLPEMLKKKAAINSFTSFCQIIINNIKKINENN
Summary
Uniprot
EMBL
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
6B3W
E-value=7.91287e-55,
Score=544
Ontologies
GO
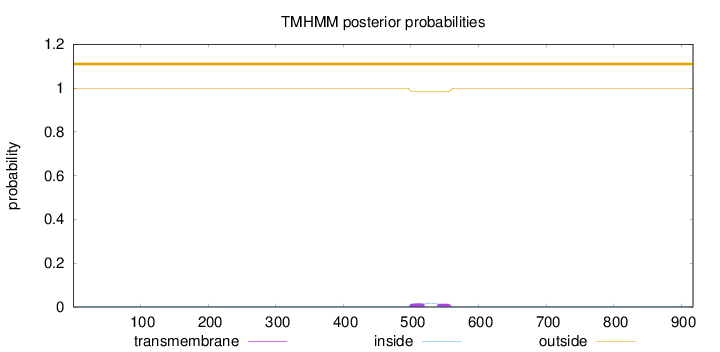
Topology
Length:
917
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.637699999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00094
outside
1 - 917
Population Genetic Test Statistics
Pi
16.703695
Theta
17.28753
Tajima's D
-0.988644
CLR
0.614065
CSRT
0.139793010349483
Interpretation
Uncertain
