Gene
KWMTBOMO06909 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004645
Annotation
suppressor_of_profilin_2_[Bombyx_mori]
Full name
Actin-related protein 2/3 complex subunit
Location in the cell
Cytoplasmic Reliability : 1.57 Extracellular Reliability : 1.859
Sequence
CDS
ATGGCACAAACGTTAACTTTTGGTGACTCATGCGCGCCGATAACATGTCATGCATGGAACAAGGATAGAAGTCAAATTGCATTCTCACCAAACAATAATGAAGTTCACATATACAAGAAGGAAGGAAATGACTGGAAGCAAACAAACAACCTTGTGGAACACGACCTTAGAGTGATGGGGATTGATTGGGCTCCCAACACCAACCGTATTGTTACGTGTTCTGTTGACCGTAATGCTTACGTCTGGACTCAAGGAGATGATGGGAAATGGGCAACTACTCTTGTTCTTCTGCGTATCAACCGAGCAGCGACTTGCGTGAAATGGTCTCCCATGGAGAACAAGTTTGCTGTCGGGTCAGGAGCTAGATTGATATCAATTTGCTATTTTGAGAAGGAAAATAATTGGTGGGTGTCTAAGCATATTAAGAAGCCCATTCGTTCGACTGTGACATCTATTGATTGGCATCCCAATAACATTTTGCTCGTCGCTGGCTCTGCTGACTTCAAAGTACGAGTCTTCTCTGCCTATATCAAGGATATCGAAGATCAGCCTGGACCGAATGTGTGGGGCTCTAAGTTGCCTTTAGGACAACTGTTAGCAGAGTTTCCCAATTCGCCTTCAGGTGGCGGTTGGGTCCACAGCGTTTCCTTCTCTGCGGACGGAAATAAAGTTGCCTGGGTCGCCCACGACAGCTCAATCAACGTAACCGACGCTACTAAAGGAAGGGCTGTCATAAAACTAAAGACTGAATACCTGCCATTCTTGGGATGCAGTTGGATCACCAACAATTCCCTGATAGTAGCAGGCCACAGCTGCATTCCGTTATTGTACTGCCACGACGGTGACGATATTAAGTATGTCGCCAAACTCGACAATACTCAGAGGAAGGAGTCTGGGGGACTGTCTGCTATGAAGAAGTTCCAGTCCTTGGATCGTCATGCTCGTATCGAGACCAGCGATACATTCTTGGACTCGATACACCAGAACGCTATCTCGTGCATCAATATTTACAAAGGAACTAAATCGCACACTAACAAGTTCAGTACTTCAGGTCTCGACGGCCAGCTCGTCATATGGGACTTGGACACTTTGGAGAGGTCATTTGAAGGAATCAAGATTGTCTAA
Protein
MAQTLTFGDSCAPITCHAWNKDRSQIAFSPNNNEVHIYKKEGNDWKQTNNLVEHDLRVMGIDWAPNTNRIVTCSVDRNAYVWTQGDDGKWATTLVLLRINRAATCVKWSPMENKFAVGSGARLISICYFEKENNWWVSKHIKKPIRSTVTSIDWHPNNILLVAGSADFKVRVFSAYIKDIEDQPGPNVWGSKLPLGQLLAEFPNSPSGGGWVHSVSFSADGNKVAWVAHDSSINVTDATKGRAVIKLKTEYLPFLGCSWITNNSLIVAGHSCIPLLYCHDGDDIKYVAKLDNTQRKESGGLSAMKKFQSLDRHARIETSDTFLDSIHQNAISCINIYKGTKSHTNKFSTSGLDGQLVIWDLDTLERSFEGIKIV
Summary
Description
Functions as component of the Arp2/3 complex which is involved in regulation of actin polymerization and together with an activating nucleation-promoting factor (NPF) mediates the formation of branched actin networks.
Similarity
Belongs to the WD repeat ARPC1 family.
Uniprot
H9J555
Q5S745
I4DR44
A0A194QPJ2
I4DJI3
H9CTT0
+ More
A0A2W1BX99 A0A2A4K6M0 W5VGT1 A0A0L7L3Y4 S4PI17 A0A212FLV7 A0A1Y1NLZ3 A0A023EQX7 A0A023ET38 A0A182GMS3 A0A1S4FGV6 A0A0K8TN71 Q171R5 A0A084VL09 A0A1Q3FXZ8 A0A182P8B0 A0A182IN19 T1DNA3 A0A2M4A469 A0A1L8E5F0 A0A182JUM4 A0A2M4BRA3 W5J1U0 A0A2M3Z0W3 A0A2M4BRI4 A0A2M3Z0Q3 A0A182F1E1 A0A182I7A7 A0A182Q2Y5 A0A182SM36 A0A182VBK3 A0A182XNP8 A0A182LMV0 Q7PWN2 A0A182U2L1 A0A182MZ76 A0A182Y0B2 U5ETL0 A0A182VUD6 A0A2J7QGZ0 A0A182RSJ6 A0A182MID2 A0A067QYD7 A0A1B0DH83 A0A1A9Z442 A0A0T6B9N5 D3TPI0 A0A1A9UYR4 A0A1A9YJV9 A0A0A1WMR5 J3JZG0 A0A1I8PPU9 A0A0L0CJN3 A0A0K8UMT1 A0A034WWR4 A0A1A9WXD7 W8C4D9 A0A1I8MC28 A0A1W4XH64 A0A1J1II70 A0A1L8ECK8 B4GX11 Q29K75 A0A336M9E9 A0A336M8T0 B3MN25 A0A1W4UV62 B4HXA3 B4Q569 O97182 B4P3Q6 B3NAD0 A0A0M5J984 B4MVR3 E2RZJ2 A0A1B6CGM3 A0A0P4VST6 A0A1B6JJC6 B4KIY1 T1HKR3 B4JCX5 A0A3B0JJK2 A0A1B6M8N3 A0A0P4WLJ6 A0A0V0GCJ0 B4M8L5 A0A069DSY9 E0VQZ3 E9HDB8 A0A3R7N738 A0A0N8E7K5 A0A0P5E8Q3 R4WDR2 A0A0L7QX55
A0A2W1BX99 A0A2A4K6M0 W5VGT1 A0A0L7L3Y4 S4PI17 A0A212FLV7 A0A1Y1NLZ3 A0A023EQX7 A0A023ET38 A0A182GMS3 A0A1S4FGV6 A0A0K8TN71 Q171R5 A0A084VL09 A0A1Q3FXZ8 A0A182P8B0 A0A182IN19 T1DNA3 A0A2M4A469 A0A1L8E5F0 A0A182JUM4 A0A2M4BRA3 W5J1U0 A0A2M3Z0W3 A0A2M4BRI4 A0A2M3Z0Q3 A0A182F1E1 A0A182I7A7 A0A182Q2Y5 A0A182SM36 A0A182VBK3 A0A182XNP8 A0A182LMV0 Q7PWN2 A0A182U2L1 A0A182MZ76 A0A182Y0B2 U5ETL0 A0A182VUD6 A0A2J7QGZ0 A0A182RSJ6 A0A182MID2 A0A067QYD7 A0A1B0DH83 A0A1A9Z442 A0A0T6B9N5 D3TPI0 A0A1A9UYR4 A0A1A9YJV9 A0A0A1WMR5 J3JZG0 A0A1I8PPU9 A0A0L0CJN3 A0A0K8UMT1 A0A034WWR4 A0A1A9WXD7 W8C4D9 A0A1I8MC28 A0A1W4XH64 A0A1J1II70 A0A1L8ECK8 B4GX11 Q29K75 A0A336M9E9 A0A336M8T0 B3MN25 A0A1W4UV62 B4HXA3 B4Q569 O97182 B4P3Q6 B3NAD0 A0A0M5J984 B4MVR3 E2RZJ2 A0A1B6CGM3 A0A0P4VST6 A0A1B6JJC6 B4KIY1 T1HKR3 B4JCX5 A0A3B0JJK2 A0A1B6M8N3 A0A0P4WLJ6 A0A0V0GCJ0 B4M8L5 A0A069DSY9 E0VQZ3 E9HDB8 A0A3R7N738 A0A0N8E7K5 A0A0P5E8Q3 R4WDR2 A0A0L7QX55
Pubmed
19121390
22651552
26354079
28756777
26227816
23622113
+ More
22118469 28004739 24945155 26483478 26369729 17510324 24438588 20920257 23761445 20966253 12364791 14747013 17210077 25244985 24845553 20353571 25830018 22516182 23537049 26108605 25348373 24495485 25315136 17994087 15632085 23185243 18057021 22936249 10886416 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27129103 26334808 20566863 21292972 23691247
22118469 28004739 24945155 26483478 26369729 17510324 24438588 20920257 23761445 20966253 12364791 14747013 17210077 25244985 24845553 20353571 25830018 22516182 23537049 26108605 25348373 24495485 25315136 17994087 15632085 23185243 18057021 22936249 10886416 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27129103 26334808 20566863 21292972 23691247
EMBL
BABH01040712
AY763110
AAV65755.1
AK404933
BAM20384.1
KQ461185
+ More
KPJ07453.1 AK401451 KQ459252 BAM18073.1 KPJ02166.1 JQ364941 ODYU01000452 AFD50557.1 SOQ35262.1 KZ149906 PZC78275.1 NWSH01000092 PCG79666.1 KF670139 AHH80647.1 JTDY01003110 KOB70130.1 GAIX01005585 JAA86975.1 AGBW02007696 OWR54722.1 GEZM01003101 JAV96917.1 GAPW01002063 JAC11535.1 GAPW01002064 JAC11534.1 JXUM01075043 KQ562865 KXJ74977.1 GDAI01002228 JAI15375.1 CH477447 EAT40752.1 ATLV01014395 KE524970 KFB38653.1 GFDL01002606 JAV32439.1 GAMD01003065 JAA98525.1 GGFK01002220 MBW35541.1 GFDF01000107 JAV13977.1 GGFJ01006474 MBW55615.1 ADMH02002157 ETN58217.1 GGFM01001370 MBW22121.1 GGFJ01006473 MBW55614.1 GGFM01001361 MBW22112.1 APCN01003375 AXCN02002069 AAAB01008984 EAA14867.2 GANO01001866 JAB58005.1 NEVH01014359 PNF27847.1 AXCM01000345 KK852879 KDR14465.1 AJVK01060758 LJIG01009020 KRT83915.1 CCAG010006778 EZ423332 ADD19608.1 GBXI01014321 JAC99970.1 APGK01055683 BT128642 KB741269 KB632330 AEE63599.1 ENN71437.1 ERL92590.1 JRES01000300 KNC32618.1 GDHF01024327 JAI27987.1 GAKP01000292 JAC58660.1 GAMC01005584 JAC00972.1 CVRI01000048 CRK98750.1 GFDG01002434 JAV16365.1 CH479195 EDW27338.1 CH379061 EAL33302.1 KRT04886.1 UFQT01000742 SSX26865.1 UFQT01000716 SSX26732.1 CH902620 EDV32003.1 KPU73731.1 CH480818 EDW51683.1 CM000361 CM002910 EDX04975.1 KMY90166.1 AE014134 AY058778 AJ133649 AAF53339.1 AAL14007.1 CAB38634.1 CM000157 EDW88357.1 CH954177 EDV58632.1 CP012523 ALC39544.1 CH963857 EDW75783.1 AB568464 BAJ23428.1 GEDC01025248 GEDC01024694 JAS12050.1 JAS12604.1 GDKW01000384 JAI56211.1 GECU01008467 JAS99239.1 CH933807 EDW13494.1 ACPB03015801 CH916368 EDW03214.1 OUUW01000006 SPP82537.1 GEBQ01007763 JAT32214.1 GDRN01037180 JAI67309.1 GECL01001021 JAP05103.1 CH940654 EDW57541.1 GBGD01001824 JAC87065.1 DS235442 EEB15799.1 GL732623 EFX70226.1 QCYY01001308 ROT78974.1 GDIQ01054028 JAN40709.1 GDIP01145135 LRGB01001899 JAJ78267.1 KZS10082.1 AK417903 BAN21118.1 KQ414705 KOC63146.1
KPJ07453.1 AK401451 KQ459252 BAM18073.1 KPJ02166.1 JQ364941 ODYU01000452 AFD50557.1 SOQ35262.1 KZ149906 PZC78275.1 NWSH01000092 PCG79666.1 KF670139 AHH80647.1 JTDY01003110 KOB70130.1 GAIX01005585 JAA86975.1 AGBW02007696 OWR54722.1 GEZM01003101 JAV96917.1 GAPW01002063 JAC11535.1 GAPW01002064 JAC11534.1 JXUM01075043 KQ562865 KXJ74977.1 GDAI01002228 JAI15375.1 CH477447 EAT40752.1 ATLV01014395 KE524970 KFB38653.1 GFDL01002606 JAV32439.1 GAMD01003065 JAA98525.1 GGFK01002220 MBW35541.1 GFDF01000107 JAV13977.1 GGFJ01006474 MBW55615.1 ADMH02002157 ETN58217.1 GGFM01001370 MBW22121.1 GGFJ01006473 MBW55614.1 GGFM01001361 MBW22112.1 APCN01003375 AXCN02002069 AAAB01008984 EAA14867.2 GANO01001866 JAB58005.1 NEVH01014359 PNF27847.1 AXCM01000345 KK852879 KDR14465.1 AJVK01060758 LJIG01009020 KRT83915.1 CCAG010006778 EZ423332 ADD19608.1 GBXI01014321 JAC99970.1 APGK01055683 BT128642 KB741269 KB632330 AEE63599.1 ENN71437.1 ERL92590.1 JRES01000300 KNC32618.1 GDHF01024327 JAI27987.1 GAKP01000292 JAC58660.1 GAMC01005584 JAC00972.1 CVRI01000048 CRK98750.1 GFDG01002434 JAV16365.1 CH479195 EDW27338.1 CH379061 EAL33302.1 KRT04886.1 UFQT01000742 SSX26865.1 UFQT01000716 SSX26732.1 CH902620 EDV32003.1 KPU73731.1 CH480818 EDW51683.1 CM000361 CM002910 EDX04975.1 KMY90166.1 AE014134 AY058778 AJ133649 AAF53339.1 AAL14007.1 CAB38634.1 CM000157 EDW88357.1 CH954177 EDV58632.1 CP012523 ALC39544.1 CH963857 EDW75783.1 AB568464 BAJ23428.1 GEDC01025248 GEDC01024694 JAS12050.1 JAS12604.1 GDKW01000384 JAI56211.1 GECU01008467 JAS99239.1 CH933807 EDW13494.1 ACPB03015801 CH916368 EDW03214.1 OUUW01000006 SPP82537.1 GEBQ01007763 JAT32214.1 GDRN01037180 JAI67309.1 GECL01001021 JAP05103.1 CH940654 EDW57541.1 GBGD01001824 JAC87065.1 DS235442 EEB15799.1 GL732623 EFX70226.1 QCYY01001308 ROT78974.1 GDIQ01054028 JAN40709.1 GDIP01145135 LRGB01001899 JAJ78267.1 KZS10082.1 AK417903 BAN21118.1 KQ414705 KOC63146.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000007151
+ More
UP000069940 UP000249989 UP000008820 UP000030765 UP000075885 UP000075880 UP000075881 UP000000673 UP000069272 UP000075840 UP000075886 UP000075901 UP000075903 UP000076407 UP000075882 UP000007062 UP000075902 UP000075884 UP000076408 UP000075920 UP000235965 UP000075900 UP000075883 UP000027135 UP000092462 UP000092445 UP000092444 UP000078200 UP000092443 UP000019118 UP000030742 UP000095300 UP000037069 UP000091820 UP000095301 UP000192223 UP000183832 UP000008744 UP000001819 UP000007801 UP000192221 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000092553 UP000007798 UP000009192 UP000015103 UP000001070 UP000268350 UP000008792 UP000009046 UP000000305 UP000283509 UP000076858 UP000053825
UP000069940 UP000249989 UP000008820 UP000030765 UP000075885 UP000075880 UP000075881 UP000000673 UP000069272 UP000075840 UP000075886 UP000075901 UP000075903 UP000076407 UP000075882 UP000007062 UP000075902 UP000075884 UP000076408 UP000075920 UP000235965 UP000075900 UP000075883 UP000027135 UP000092462 UP000092445 UP000092444 UP000078200 UP000092443 UP000019118 UP000030742 UP000095300 UP000037069 UP000091820 UP000095301 UP000192223 UP000183832 UP000008744 UP000001819 UP000007801 UP000192221 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000092553 UP000007798 UP000009192 UP000015103 UP000001070 UP000268350 UP000008792 UP000009046 UP000000305 UP000283509 UP000076858 UP000053825
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9J555
Q5S745
I4DR44
A0A194QPJ2
I4DJI3
H9CTT0
+ More
A0A2W1BX99 A0A2A4K6M0 W5VGT1 A0A0L7L3Y4 S4PI17 A0A212FLV7 A0A1Y1NLZ3 A0A023EQX7 A0A023ET38 A0A182GMS3 A0A1S4FGV6 A0A0K8TN71 Q171R5 A0A084VL09 A0A1Q3FXZ8 A0A182P8B0 A0A182IN19 T1DNA3 A0A2M4A469 A0A1L8E5F0 A0A182JUM4 A0A2M4BRA3 W5J1U0 A0A2M3Z0W3 A0A2M4BRI4 A0A2M3Z0Q3 A0A182F1E1 A0A182I7A7 A0A182Q2Y5 A0A182SM36 A0A182VBK3 A0A182XNP8 A0A182LMV0 Q7PWN2 A0A182U2L1 A0A182MZ76 A0A182Y0B2 U5ETL0 A0A182VUD6 A0A2J7QGZ0 A0A182RSJ6 A0A182MID2 A0A067QYD7 A0A1B0DH83 A0A1A9Z442 A0A0T6B9N5 D3TPI0 A0A1A9UYR4 A0A1A9YJV9 A0A0A1WMR5 J3JZG0 A0A1I8PPU9 A0A0L0CJN3 A0A0K8UMT1 A0A034WWR4 A0A1A9WXD7 W8C4D9 A0A1I8MC28 A0A1W4XH64 A0A1J1II70 A0A1L8ECK8 B4GX11 Q29K75 A0A336M9E9 A0A336M8T0 B3MN25 A0A1W4UV62 B4HXA3 B4Q569 O97182 B4P3Q6 B3NAD0 A0A0M5J984 B4MVR3 E2RZJ2 A0A1B6CGM3 A0A0P4VST6 A0A1B6JJC6 B4KIY1 T1HKR3 B4JCX5 A0A3B0JJK2 A0A1B6M8N3 A0A0P4WLJ6 A0A0V0GCJ0 B4M8L5 A0A069DSY9 E0VQZ3 E9HDB8 A0A3R7N738 A0A0N8E7K5 A0A0P5E8Q3 R4WDR2 A0A0L7QX55
A0A2W1BX99 A0A2A4K6M0 W5VGT1 A0A0L7L3Y4 S4PI17 A0A212FLV7 A0A1Y1NLZ3 A0A023EQX7 A0A023ET38 A0A182GMS3 A0A1S4FGV6 A0A0K8TN71 Q171R5 A0A084VL09 A0A1Q3FXZ8 A0A182P8B0 A0A182IN19 T1DNA3 A0A2M4A469 A0A1L8E5F0 A0A182JUM4 A0A2M4BRA3 W5J1U0 A0A2M3Z0W3 A0A2M4BRI4 A0A2M3Z0Q3 A0A182F1E1 A0A182I7A7 A0A182Q2Y5 A0A182SM36 A0A182VBK3 A0A182XNP8 A0A182LMV0 Q7PWN2 A0A182U2L1 A0A182MZ76 A0A182Y0B2 U5ETL0 A0A182VUD6 A0A2J7QGZ0 A0A182RSJ6 A0A182MID2 A0A067QYD7 A0A1B0DH83 A0A1A9Z442 A0A0T6B9N5 D3TPI0 A0A1A9UYR4 A0A1A9YJV9 A0A0A1WMR5 J3JZG0 A0A1I8PPU9 A0A0L0CJN3 A0A0K8UMT1 A0A034WWR4 A0A1A9WXD7 W8C4D9 A0A1I8MC28 A0A1W4XH64 A0A1J1II70 A0A1L8ECK8 B4GX11 Q29K75 A0A336M9E9 A0A336M8T0 B3MN25 A0A1W4UV62 B4HXA3 B4Q569 O97182 B4P3Q6 B3NAD0 A0A0M5J984 B4MVR3 E2RZJ2 A0A1B6CGM3 A0A0P4VST6 A0A1B6JJC6 B4KIY1 T1HKR3 B4JCX5 A0A3B0JJK2 A0A1B6M8N3 A0A0P4WLJ6 A0A0V0GCJ0 B4M8L5 A0A069DSY9 E0VQZ3 E9HDB8 A0A3R7N738 A0A0N8E7K5 A0A0P5E8Q3 R4WDR2 A0A0L7QX55
PDB
6DEC
E-value=3.03199e-118,
Score=1087
Ontologies
GO
PANTHER
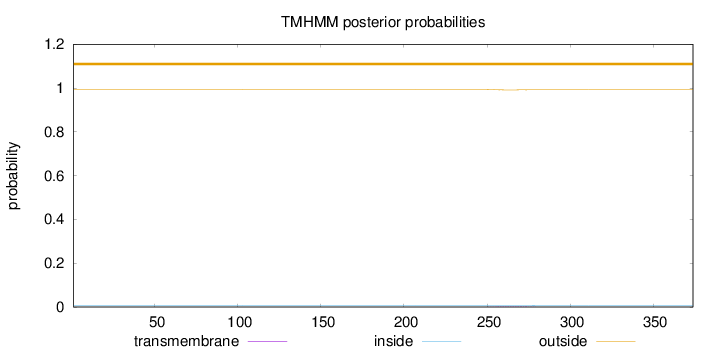
Topology
Length:
374
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03944
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00704
outside
1 - 374
Population Genetic Test Statistics
Pi
28.90864
Theta
25.970824
Tajima's D
0.494045
CLR
1.571118
CSRT
0.517674116294185
Interpretation
Uncertain
