Pre Gene Modal
BGIBMGA004646
Annotation
PREDICTED:_mitotic_checkpoint_protein_BUB3_[Papilio_machaon]
Full name
Mitotic checkpoint protein BUB3
Location in the cell
Nuclear Reliability : 1.564
Sequence
CDS
ATGTCAGCACAGACTAAGAAAATTGCTTCCACAATGACTGTCACACGGGTGGCGGAGTCGCGCACCGAGTTCAAATTAAAAAGCTTACCCGAAGACGCTATTTCGAGCGTTAAATTTGCACCGAAATCTAACCAGTACTTGCTGGTATCTTCATGGGACTGTTCTGTGAGGCTTTATGATGTGACCGCGAATATAGAAAGACATAAATACAATCATGAACTGCCGGTTCTTGATGTTTGTTTTCGGGATGCTGTTCATTCATATAGTGGTGGACTAGATCAGACTCTCAAAATGTATGATTTGAATGCTAGCACGGAGACTATCCTCGGAGAGCACAAAGGAGCTATACGCTGTGTAGAGTTCGCATCTGAACTTAATGCAGTACTCACAGGAAGCTGGGATGGAACAGTCAAAATGTGGGATAGCAGAGTTCCAAACTGTGTCGGCACTTACAATCAGGGAAATGAACGGGTATACACAATGAGCATAGTTGGCGAAAAGTTCGTAGTTGGTACATCGGGACGTAAAATATTTGTATGGGACGTTCGTAACATGGGGCATGTAAATCAAAGGAGGGAATCCTCACTCAAATATCAAACACGATGCATACGAGTGTTTCCCAATAAGCAGGGCTACGTACTCAGCTCAATAGAGGGACGTGTGGCGGTGGAGTATTTGGACAGTAATCCTGAGGTGCAGAAGAAGAAATATGCTTTCAAATGCCATCGGATTAAAGAAGGAGGTCTAGAAAAGATATATCCAGTGAACGCGATCAGCTTCCACTCCGTGTACAACACGTTCGCGACGGGCGGCTCGGACGGCTACGTGAACATCTGGGACGGCTTCAACAAGAAGCGGCTGTGCCAGTTCCACCGCTACAACACCGCCGTCTCCGCGCTCGCCTTCTCGCACGACGGCTCCGCGCTCGCCATCGCCTGCTCGCAGCTCGACGACTCGCAGATCGAAGCCTCCAAGCCCGAAGACACCATCTACATACGATACGTCACGGACCAGGAAACGAAGCCTAAATGA
Protein
MSAQTKKIASTMTVTRVAESRTEFKLKSLPEDAISSVKFAPKSNQYLLVSSWDCSVRLYDVTANIERHKYNHELPVLDVCFRDAVHSYSGGLDQTLKMYDLNASTETILGEHKGAIRCVEFASELNAVLTGSWDGTVKMWDSRVPNCVGTYNQGNERVYTMSIVGEKFVVGTSGRKIFVWDVRNMGHVNQRRESSLKYQTRCIRVFPNKQGYVLSSIEGRVAVEYLDSNPEVQKKKYAFKCHRIKEGGLEKIYPVNAISFHSVYNTFATGGSDGYVNIWDGFNKKRLCQFHRYNTAVSALAFSHDGSALAIACSQLDDSQIEASKPEDTIYIRYVTDQETKPK
Summary
Description
Has a dual function in spindle-assembly checkpoint signaling and in promoting the establishment of correct kinetochore-microtubule (K-MT) attachments. Promotes the formation of stable end-on bipolar attachments. Necessary for kinetochore localization of BUB1. Regulates chromosome segregation during oocyte meiosis. The BUB1/BUB3 complex plays a role in the inhibition of anaphase-promoting complex or cyclosome (APC/C) when spindle-assembly checkpoint is activated and inhibits the ubiquitin ligase activity of APC/C by phosphorylating its activator CDC20. This complex can also phosphorylate MAD1L1.
Subunit
Interacts with BUB1 and BUBR1. The BUB1/BUB3 complex interacts with MAD1L1. Interacts with ZNF207/BuGZ; leading to promote stability and kinetochore loading of BUB3.
Similarity
Belongs to the WD repeat BUB3 family.
Keywords
Acetylation
ADP-ribosylation
Alternative splicing
Cell cycle
Cell division
Centromere
Chromosome
Chromosome partition
Complete proteome
Isopeptide bond
Kinetochore
Meiosis
Mitosis
Nucleus
Phosphoprotein
Reference proteome
Repeat
Ubl conjugation
WD repeat
Feature
chain Mitotic checkpoint protein BUB3
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J556
A0A194QR56
A0A2H1V339
S4P5H9
A0A194Q9S5
A0A2W1C2C1
+ More
A0A2A4K7N2 A0A3S2NQT9 A0A212FLV0 A0A0L7L480 A0A2J7PN74 A0A067RB95 A0A1B6EGI1 A0A1B6FJ92 A0A1B6HME9 A0A1B6KU11 A0A1B6MF64 D6X4M0 A0A1W4W2W7 J9K808 A0A1Y1K2K2 A0A2T7P896 A0A1S3IJW8 A0A0A9WLL1 C3Y5S4 A0A0B6Y9E7 V4B165 R7USZ3 A0A195CU12 A0A0N7ZCD8 E2AWS0 A0A0L7RK02 A0A154NYT1 A0A195BB82 E2BFL7 A0A151J4R0 F4WDL5 A0A151JV88 A0A158NIM9 A0A2C9JQ13 A0A0J7L279 A0A026WZW0 A0A310SEH1 A0A151WQ25 A0A3R7SJK4 C0IRC6 A0A2A3EIJ3 A0A088AQL5 A0A232F546 K7IMF7 A0A1S3DC81 A0A3Q0J6B9 A0A293M4M7 A0A210PP70 A0A0L8FLL0 A0A2P2I2J4 K1PHV7 A0A3S1C938 J3JUZ6 S4RSZ5 T1IIQ6 A0A087UDV1 W5MNG7 W4Y561 R0LED4 V9KPJ9 W5K9X2 A0A1S2ZI75 K7FM27 A0A2I0MEU7 A0A1V4KJU8 A0A2I0MES4 U3J5P9 A0A3Q0GU34 A0A3B4BDS9 A0A3Q0GPJ4 A0A218UT65 U3JQ70 A0A3S2P801 H2M0R6 A0A3P9IXW3 G3W9J5 G3W9J6 A0A1L8FEC6 G1NGF8 G3W9J7 O43684-2 A0A1U7R8K2 A0A286XDC6 Q4FZS2 A0A384BM44 K9IJA3 A0A341BXJ2 F7B951 A0A3Q2HZV6 A0A2U4BRA8 A0A340WWE3 A0A2I3NAM9 A0A2K5EN33 A0A2Y9SFT9
A0A2A4K7N2 A0A3S2NQT9 A0A212FLV0 A0A0L7L480 A0A2J7PN74 A0A067RB95 A0A1B6EGI1 A0A1B6FJ92 A0A1B6HME9 A0A1B6KU11 A0A1B6MF64 D6X4M0 A0A1W4W2W7 J9K808 A0A1Y1K2K2 A0A2T7P896 A0A1S3IJW8 A0A0A9WLL1 C3Y5S4 A0A0B6Y9E7 V4B165 R7USZ3 A0A195CU12 A0A0N7ZCD8 E2AWS0 A0A0L7RK02 A0A154NYT1 A0A195BB82 E2BFL7 A0A151J4R0 F4WDL5 A0A151JV88 A0A158NIM9 A0A2C9JQ13 A0A0J7L279 A0A026WZW0 A0A310SEH1 A0A151WQ25 A0A3R7SJK4 C0IRC6 A0A2A3EIJ3 A0A088AQL5 A0A232F546 K7IMF7 A0A1S3DC81 A0A3Q0J6B9 A0A293M4M7 A0A210PP70 A0A0L8FLL0 A0A2P2I2J4 K1PHV7 A0A3S1C938 J3JUZ6 S4RSZ5 T1IIQ6 A0A087UDV1 W5MNG7 W4Y561 R0LED4 V9KPJ9 W5K9X2 A0A1S2ZI75 K7FM27 A0A2I0MEU7 A0A1V4KJU8 A0A2I0MES4 U3J5P9 A0A3Q0GU34 A0A3B4BDS9 A0A3Q0GPJ4 A0A218UT65 U3JQ70 A0A3S2P801 H2M0R6 A0A3P9IXW3 G3W9J5 G3W9J6 A0A1L8FEC6 G1NGF8 G3W9J7 O43684-2 A0A1U7R8K2 A0A286XDC6 Q4FZS2 A0A384BM44 K9IJA3 A0A341BXJ2 F7B951 A0A3Q2HZV6 A0A2U4BRA8 A0A340WWE3 A0A2I3NAM9 A0A2K5EN33 A0A2Y9SFT9
Pubmed
19121390
26354079
23622113
28756777
22118469
26227816
+ More
24845553 18362917 19820115 28004739 25401762 26823975 18563158 23254933 20798317 21719571 21347285 15562597 24508170 30249741 28648823 20075255 28812685 22992520 22516182 23537049 24402279 25329095 17381049 23371554 23749191 25463417 17554307 21709235 27762356 20838655 10198256 9660858 14702039 15164054 15489334 15525512 18199686 18669648 19608861 21269460 23186163 24462186 24462187 21993624 15057822 15632090 24813606 19892987
24845553 18362917 19820115 28004739 25401762 26823975 18563158 23254933 20798317 21719571 21347285 15562597 24508170 30249741 28648823 20075255 28812685 22992520 22516182 23537049 24402279 25329095 17381049 23371554 23749191 25463417 17554307 21709235 27762356 20838655 10198256 9660858 14702039 15164054 15489334 15525512 18199686 18669648 19608861 21269460 23186163 24462186 24462187 21993624 15057822 15632090 24813606 19892987
EMBL
BABH01040711
KQ461185
KPJ07455.1
ODYU01000452
SOQ35260.1
GAIX01005539
+ More
JAA87021.1 KQ459252 KPJ02164.1 KZ149906 PZC78273.1 NWSH01000092 PCG79662.1 RSAL01000138 RVE46176.1 AGBW02007696 OWR54724.1 JTDY01003110 KOB70131.1 NEVH01023957 PNF17793.1 KK852811 KDR15933.1 GEDC01003709 GEDC01000297 JAS33589.1 JAS37001.1 GECZ01019522 JAS50247.1 GECU01031872 JAS75834.1 GEBQ01025045 JAT14932.1 GEBQ01005379 JAT34598.1 KQ971380 EEZ97704.1 ABLF02028424 GEZM01098963 JAV53606.1 PZQS01000005 PVD29616.1 GBHO01036206 GBRD01008691 GDHC01016182 JAG07398.1 JAG57130.1 JAQ02447.1 GG666487 EEN64321.1 HACG01005566 CEK52431.1 KB204089 ESO81944.1 AMQN01006420 AMQN01006421 AMQN01006422 KB298315 ELU09330.1 KQ977306 KYN03624.1 GDRN01068652 JAI64168.1 GL443424 EFN62106.1 KQ414578 KOC71195.1 KQ434777 KZC04234.1 KQ976529 KYM81811.1 GL448037 EFN85539.1 KQ980102 KYN17747.1 GL888087 EGI67795.1 KQ981702 KYN37459.1 ADTU01017000 LBMM01001164 KMQ96796.1 KK107072 QOIP01000007 EZA60659.1 RLU20878.1 KQ762400 OAD55898.1 KQ982850 KYQ49966.1 QCYY01003397 ROT63696.1 EU492542 ACD13595.1 KZ288253 PBC31006.1 NNAY01000987 OXU25600.1 GFWV01009206 MAA33935.1 NEDP02005569 OWF38295.1 KQ429209 KOF65543.1 IACF01002573 LAB68221.1 JH816779 EKC21138.1 RQTK01000143 RUS86307.1 BT127061 KB632379 AEE62023.1 ERL94090.1 JH430212 KK119382 KFM75540.1 AHAT01004030 AAGJ04038112 KB743453 EOA98647.1 JW867677 AFP00195.1 AGCU01172420 AGCU01172421 AKCR02000017 PKK28182.1 LSYS01003057 OPJ84097.1 PKK28181.1 ADON01095586 MUZQ01000141 OWK56963.1 AGTO01011741 CM012455 RVE59742.1 AEFK01004272 AEFK01004273 CM004479 OCT69916.1 AF047472 AF047473 AF053304 AF081496 AK312546 AC012391 CH471066 BC005138 BC022438 AAKN02002977 AABR07005873 BC099199 CH473953 AAH99199.1 EDM11706.1 GABZ01006998 JAA46527.1 AHZZ02036186
JAA87021.1 KQ459252 KPJ02164.1 KZ149906 PZC78273.1 NWSH01000092 PCG79662.1 RSAL01000138 RVE46176.1 AGBW02007696 OWR54724.1 JTDY01003110 KOB70131.1 NEVH01023957 PNF17793.1 KK852811 KDR15933.1 GEDC01003709 GEDC01000297 JAS33589.1 JAS37001.1 GECZ01019522 JAS50247.1 GECU01031872 JAS75834.1 GEBQ01025045 JAT14932.1 GEBQ01005379 JAT34598.1 KQ971380 EEZ97704.1 ABLF02028424 GEZM01098963 JAV53606.1 PZQS01000005 PVD29616.1 GBHO01036206 GBRD01008691 GDHC01016182 JAG07398.1 JAG57130.1 JAQ02447.1 GG666487 EEN64321.1 HACG01005566 CEK52431.1 KB204089 ESO81944.1 AMQN01006420 AMQN01006421 AMQN01006422 KB298315 ELU09330.1 KQ977306 KYN03624.1 GDRN01068652 JAI64168.1 GL443424 EFN62106.1 KQ414578 KOC71195.1 KQ434777 KZC04234.1 KQ976529 KYM81811.1 GL448037 EFN85539.1 KQ980102 KYN17747.1 GL888087 EGI67795.1 KQ981702 KYN37459.1 ADTU01017000 LBMM01001164 KMQ96796.1 KK107072 QOIP01000007 EZA60659.1 RLU20878.1 KQ762400 OAD55898.1 KQ982850 KYQ49966.1 QCYY01003397 ROT63696.1 EU492542 ACD13595.1 KZ288253 PBC31006.1 NNAY01000987 OXU25600.1 GFWV01009206 MAA33935.1 NEDP02005569 OWF38295.1 KQ429209 KOF65543.1 IACF01002573 LAB68221.1 JH816779 EKC21138.1 RQTK01000143 RUS86307.1 BT127061 KB632379 AEE62023.1 ERL94090.1 JH430212 KK119382 KFM75540.1 AHAT01004030 AAGJ04038112 KB743453 EOA98647.1 JW867677 AFP00195.1 AGCU01172420 AGCU01172421 AKCR02000017 PKK28182.1 LSYS01003057 OPJ84097.1 PKK28181.1 ADON01095586 MUZQ01000141 OWK56963.1 AGTO01011741 CM012455 RVE59742.1 AEFK01004272 AEFK01004273 CM004479 OCT69916.1 AF047472 AF047473 AF053304 AF081496 AK312546 AC012391 CH471066 BC005138 BC022438 AAKN02002977 AABR07005873 BC099199 CH473953 AAH99199.1 EDM11706.1 GABZ01006998 JAA46527.1 AHZZ02036186
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000007151
+ More
UP000037510 UP000235965 UP000027135 UP000007266 UP000192223 UP000007819 UP000245119 UP000085678 UP000001554 UP000030746 UP000014760 UP000078542 UP000000311 UP000053825 UP000076502 UP000078540 UP000008237 UP000078492 UP000007755 UP000078541 UP000005205 UP000076420 UP000036403 UP000053097 UP000279307 UP000075809 UP000283509 UP000242457 UP000005203 UP000215335 UP000002358 UP000079169 UP000242188 UP000053454 UP000005408 UP000271974 UP000030742 UP000245300 UP000054359 UP000018468 UP000007110 UP000018467 UP000079721 UP000007267 UP000053872 UP000190648 UP000016666 UP000189705 UP000261520 UP000197619 UP000016665 UP000001038 UP000265180 UP000265200 UP000007648 UP000186698 UP000001645 UP000005640 UP000189706 UP000005447 UP000002494 UP000261680 UP000291021 UP000252040 UP000002281 UP000245320 UP000265300 UP000028761 UP000233020 UP000248484
UP000037510 UP000235965 UP000027135 UP000007266 UP000192223 UP000007819 UP000245119 UP000085678 UP000001554 UP000030746 UP000014760 UP000078542 UP000000311 UP000053825 UP000076502 UP000078540 UP000008237 UP000078492 UP000007755 UP000078541 UP000005205 UP000076420 UP000036403 UP000053097 UP000279307 UP000075809 UP000283509 UP000242457 UP000005203 UP000215335 UP000002358 UP000079169 UP000242188 UP000053454 UP000005408 UP000271974 UP000030742 UP000245300 UP000054359 UP000018468 UP000007110 UP000018467 UP000079721 UP000007267 UP000053872 UP000190648 UP000016666 UP000189705 UP000261520 UP000197619 UP000016665 UP000001038 UP000265180 UP000265200 UP000007648 UP000186698 UP000001645 UP000005640 UP000189706 UP000005447 UP000002494 UP000261680 UP000291021 UP000252040 UP000002281 UP000245320 UP000265300 UP000028761 UP000233020 UP000248484
Pfam
PF00400 WD40
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9J556
A0A194QR56
A0A2H1V339
S4P5H9
A0A194Q9S5
A0A2W1C2C1
+ More
A0A2A4K7N2 A0A3S2NQT9 A0A212FLV0 A0A0L7L480 A0A2J7PN74 A0A067RB95 A0A1B6EGI1 A0A1B6FJ92 A0A1B6HME9 A0A1B6KU11 A0A1B6MF64 D6X4M0 A0A1W4W2W7 J9K808 A0A1Y1K2K2 A0A2T7P896 A0A1S3IJW8 A0A0A9WLL1 C3Y5S4 A0A0B6Y9E7 V4B165 R7USZ3 A0A195CU12 A0A0N7ZCD8 E2AWS0 A0A0L7RK02 A0A154NYT1 A0A195BB82 E2BFL7 A0A151J4R0 F4WDL5 A0A151JV88 A0A158NIM9 A0A2C9JQ13 A0A0J7L279 A0A026WZW0 A0A310SEH1 A0A151WQ25 A0A3R7SJK4 C0IRC6 A0A2A3EIJ3 A0A088AQL5 A0A232F546 K7IMF7 A0A1S3DC81 A0A3Q0J6B9 A0A293M4M7 A0A210PP70 A0A0L8FLL0 A0A2P2I2J4 K1PHV7 A0A3S1C938 J3JUZ6 S4RSZ5 T1IIQ6 A0A087UDV1 W5MNG7 W4Y561 R0LED4 V9KPJ9 W5K9X2 A0A1S2ZI75 K7FM27 A0A2I0MEU7 A0A1V4KJU8 A0A2I0MES4 U3J5P9 A0A3Q0GU34 A0A3B4BDS9 A0A3Q0GPJ4 A0A218UT65 U3JQ70 A0A3S2P801 H2M0R6 A0A3P9IXW3 G3W9J5 G3W9J6 A0A1L8FEC6 G1NGF8 G3W9J7 O43684-2 A0A1U7R8K2 A0A286XDC6 Q4FZS2 A0A384BM44 K9IJA3 A0A341BXJ2 F7B951 A0A3Q2HZV6 A0A2U4BRA8 A0A340WWE3 A0A2I3NAM9 A0A2K5EN33 A0A2Y9SFT9
A0A2A4K7N2 A0A3S2NQT9 A0A212FLV0 A0A0L7L480 A0A2J7PN74 A0A067RB95 A0A1B6EGI1 A0A1B6FJ92 A0A1B6HME9 A0A1B6KU11 A0A1B6MF64 D6X4M0 A0A1W4W2W7 J9K808 A0A1Y1K2K2 A0A2T7P896 A0A1S3IJW8 A0A0A9WLL1 C3Y5S4 A0A0B6Y9E7 V4B165 R7USZ3 A0A195CU12 A0A0N7ZCD8 E2AWS0 A0A0L7RK02 A0A154NYT1 A0A195BB82 E2BFL7 A0A151J4R0 F4WDL5 A0A151JV88 A0A158NIM9 A0A2C9JQ13 A0A0J7L279 A0A026WZW0 A0A310SEH1 A0A151WQ25 A0A3R7SJK4 C0IRC6 A0A2A3EIJ3 A0A088AQL5 A0A232F546 K7IMF7 A0A1S3DC81 A0A3Q0J6B9 A0A293M4M7 A0A210PP70 A0A0L8FLL0 A0A2P2I2J4 K1PHV7 A0A3S1C938 J3JUZ6 S4RSZ5 T1IIQ6 A0A087UDV1 W5MNG7 W4Y561 R0LED4 V9KPJ9 W5K9X2 A0A1S2ZI75 K7FM27 A0A2I0MEU7 A0A1V4KJU8 A0A2I0MES4 U3J5P9 A0A3Q0GU34 A0A3B4BDS9 A0A3Q0GPJ4 A0A218UT65 U3JQ70 A0A3S2P801 H2M0R6 A0A3P9IXW3 G3W9J5 G3W9J6 A0A1L8FEC6 G1NGF8 G3W9J7 O43684-2 A0A1U7R8K2 A0A286XDC6 Q4FZS2 A0A384BM44 K9IJA3 A0A341BXJ2 F7B951 A0A3Q2HZV6 A0A2U4BRA8 A0A340WWE3 A0A2I3NAM9 A0A2K5EN33 A0A2Y9SFT9
PDB
4OWR
E-value=1.77641e-45,
Score=459
Ontologies
GO
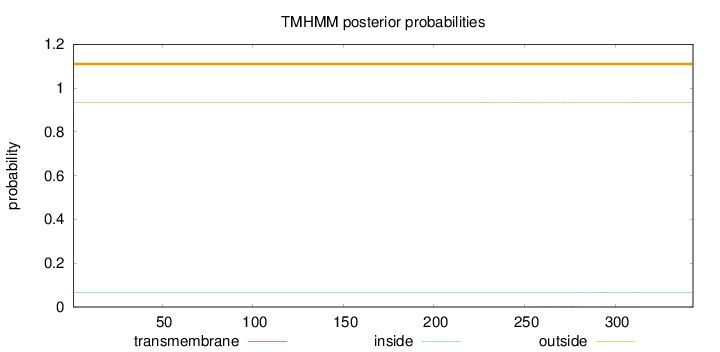
Topology
Subcellular location
Nucleus
Starts to localize at kinetochores in prometaphase I (Pro-MI) stage and maintains the localization until the metaphase I-anaphase I (MI-AI) transition. With evidence from 4 publications.
Chromosome Starts to localize at kinetochores in prometaphase I (Pro-MI) stage and maintains the localization until the metaphase I-anaphase I (MI-AI) transition. With evidence from 4 publications.
Centromere Starts to localize at kinetochores in prometaphase I (Pro-MI) stage and maintains the localization until the metaphase I-anaphase I (MI-AI) transition. With evidence from 4 publications.
Kinetochore Starts to localize at kinetochores in prometaphase I (Pro-MI) stage and maintains the localization until the metaphase I-anaphase I (MI-AI) transition. With evidence from 4 publications.
Chromosome Starts to localize at kinetochores in prometaphase I (Pro-MI) stage and maintains the localization until the metaphase I-anaphase I (MI-AI) transition. With evidence from 4 publications.
Centromere Starts to localize at kinetochores in prometaphase I (Pro-MI) stage and maintains the localization until the metaphase I-anaphase I (MI-AI) transition. With evidence from 4 publications.
Kinetochore Starts to localize at kinetochores in prometaphase I (Pro-MI) stage and maintains the localization until the metaphase I-anaphase I (MI-AI) transition. With evidence from 4 publications.
Length:
343
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05587
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.06641
outside
1 - 343
Population Genetic Test Statistics
Pi
16.427683
Theta
17.653741
Tajima's D
-0.316306
CLR
12.487709
CSRT
0.295285235738213
Interpretation
Uncertain
