Gene
KWMTBOMO06900
Pre Gene Modal
BGIBMGA007548
Annotation
retroelement_polyprotein_[Glyptapanteles_flavicoxis]
Location in the cell
Nuclear Reliability : 3.914
Sequence
CDS
ATGAGTGAGCCTTGTGATGGAGTTGGTGGCGTCAGTTTATCTGACGTCACCTTTTCCAAAAATTCTCCGGAGGTCGGTGCGCCGTCATCAGAAGTGGGATCACCTCCCACTTCTGCTCGCCCTTCTGGCTCGGTTAACAAAAATATGAACACTTCTGTTGATGGGGTGCCGGCTGATTTTTTCACTCAGATGTTGCAAATGATGAAACACGTGTCGGATCGTATGACGTCGCAATCAGATATTTCAAAAGTGAGAATAAATGACGTGTTTTTGCCGTCATACGATCCAGACGCTAATGTTGGGGTTAGAGAATGGTGTCAACATGTGACCATAGCTATGGAAACTTACAACCTGAGCGATTATGAAGTCCGGATGAAGGCTGGTAGTTTGCTTAAGGGTCGCGCAAGGTTGTGGGTTGACAATTGGCTGGTGTCCACCACCACGTGGCAAGAATTGCGTGATGTTTTAATTTCGACTTTTGAACCGGAAAACCGATATTCACGCGATGTTGTGCGTTTTAGAGAACATATTTACGATAATTCTAAAGACATCGCTCAGTTCTTATCACAAGCTTGGGTTTTATGGAGACGTATTACTAAAGATAAGTTGTCGAATGATGACGCGGTTGAGGCTGTTATCGGTTGCATTGGGGATGAGCGCCTGAGAATCGAATTGTTAAATGCGCGAGCTACATCTGTCCCAGAACTTATTTCAGTGGCCTCATCTATAAAAAGTGTTAAACGTCCTCATCCCAATGCATCGATACCGCATCAGAGTGTCGCAAAGCGCCCACGTTTTGATGAGACACCGGGCTTTTGCCAAATCTGTAAAAGGTCGGGTCACGATACGCGTGATTGCCGATACCACGACAAACCTGAAGGCCCTCAGCCGCGACAACAGGATGGTAAAACTACTCCTCCAGGTGTTGGTAGACCGACATGTACGTTTTGTTCAAGGTTGGGTCATACTTATCAAACGTGTTACAAGCGAGAACGTGCAGTTGTTTCTAATGTCAATTGTGTGGGGACGTCAAAATTAAATGCGATGCCAGTTGTTGTAGGAGGTTTCAAAATTAATGCAATTTTTGATAGTGGAGCCGAATGTTCAGTCATTCGTGAATCTGTAGCTGCAAAGTTGCCGGGAAAACGCGTTGATAGGGTTAGCTATTTGAAAGCTCTCGGTCAATTTACGGTTGTCTCGCTTTCAACATTAACAACAGTTTGCATAATCGACAACTTGCGCGTAGAACTAGAGTTTCACGTCGTTTCCGATATTGAAGTGAGCTCTGATATTTTGATAGGCATTAATTTGATAGAAAATACGAACCTAAGCGTTGTTGTAAATTCACGAGGTGCAATGCTGGTTCATCAACCGATCATTTACCACATGCGATCTCAAAGCCCAAAATTTGGCAAATTAGATTGTGACTTAATCAATGACAATCAGATTAGCGAGTTAAAAATATTGCTGAACAAGTTCGAACACTTGTTTATTCATGGCTACCCGCATACGCGAGTTAATACGGGTGAGCTAGAGATTCGATTAAAGGACGAAAGTAAGACAGTGGAGCGTAGGCCCTATCGGCTTAGTCCAGTTGAACGGGAAAAAGTTCGCGATATCGTTAACGAATTGCTTGAGCATAAAATAATTCGGGAAAGTAAATCTCCTTTTGCAAGCCCAATAATTATAGTTAAAAAGAAAAACGGCAAAGATCGTATGTGTGTAGACTACCGAGAGTTAAATAGGAATACCGTCAGGGACCATTATCCCTTACCGATAATCTCAGACCAAATAGACCAACTGGCCGGGGGTATTTATTTCACCACAATTGACATGGCAGCTGGGTTTCATCAGATACCTATTTCGGAGGGATCAATTGAAAAGACTGCTTTTGTAACTCCCGATGGTTTATATGAATACCTAACTATGCCATTTGGACTTTGCAACGCGGTGTCAGTCTACCAGCGCTGCATTAATAGGGCTCTCGCTCATCTCACGAGCTCACCAGACCAAGTGTGCCAGGTCTATGTCGATGATGTGCTCTCAAAGTGTAAAGATTTTGATCAGGGCTTATCCCATATTGAACGCATATTGATTGCTCTACAAGAAGCTGGCTTTTCTATTAACGTTGATAAAACCGCCTTCTTTAAACGCTCCATCGAGTACTTGGGCAATATAATTGCCGACGGGCAGGTCAGCCCTAGCCCAAGAAAGGTGGAGGCTTTAACGAAGGCACCAATCCCGAAAACTGTCAAACAAGTTAGACAATTCAATGGCCTGGCTGGATATTTCAGACGTTTCATTCCCAACTTCTCGCGAGTTATGGCGCCTTTATATGAACTCACTAAGAACGATACGACATGGGAATGGAACGAAAGGCATGACGAAGCTAGGGATTATATTGTCAAACATCTGTCTACTGCGCCTACCCTTACGCTATTCCAGGAAGACGCACCCATCGAACTCTACACGGACGCCAGCAGCATCGGCTACGGGGCAGTGTTAGTGCAGACATTCGCGGGTCGCCAACATCCAGTCGCGTACATGAGCCAGCGCACCACCGACGCCGAGAGCCGCTACCACTCTTACGAACTTGAAACGTTGGCGGTCGTAAGAGCAATTAAGCATTTTCGGCATTACCTCTACGGCCGAAAATTCAAGGTAATAACCGACTGTAATGCTCTCAAGGCCTCCAAAAACAAAAAGGACTTGCTTCCGCGTATCCATCGGTGGTGGGCGTTTTTGCAAAACTACGAATTCGAGGTGGAGTATAGGAAAGGTGAGCGATTACAGCACGCAGACTTTTTCAGTAGAAACCCTTGCAGCGAGATGTCAGTTAACGTTATGACTCGAGATATGGAATGGCTGCAGATTGAGCAGCACCGCGATGACGTGCTACGCCCAATGATGGAGAGTATAACATCTGGCGATTCCATCGAAGGCTATGTCATGGAAGATGGCATACTTAAGAAAATGCTCACAGATCCTATACTCGGCATACATAAACCAATTGTAGTGCCTAAGTCTTTCCAGTGGAGCTTGATAAACTCCTTCCACGTGACACTGCAGCACCCAGGATGGGAGAAAACTCTGCAGAAGCTACGAGAAACGTACTGGTTTGACAAGATGAACTCGGCTGTTAGACGTTTTGTAGACCATTGTGTTATATGTAGAACCTCGAAAGGACCATCTGGTTCTATACAAGCGCAGCTCCATCCGATCCAGAAACCCACGGCAGCGTTCCAGGTCATTCATATGGACATCACAGGAAAATTGGGCACGAGAAACTCCGAAGGACAATATGAGTACGTAATTGTTGCTATTGACGCATTCACGAAATACATATTACTGAGCTATTCCAATGACAAGAGCCCGAGCAGTACACTCGCGGCACTCAAGCGTGTCATTCACCTGTTTGGCACCCCGGTACAAGTGGTGGTTGATGGCGGTCGGGAGTTTCTCGGCGAGTTCAAAGTCTACACTGACCGCATAGGGATTGACGTGCATTCAACTGCGCCAGGAACAAGCCGAGCAAACGGACAGGTCGAAAGGGTCATGGGCACATTGAAAAATGCCCTTACCATGATCAAAAACTACAGTACTCAAGACTGGCAGACGGCGCTGGAAGCTTTACAGCTCGCTTTCAACTGCACGCCACATCGAGTGACAGGAGTAGCTCCGCTCACTCTCCTGACCCGCCGACAACATTGCATACCGCCAGAGTTACTAAGGTTGGTGAATATTGAGAACGAATGCATAGATTTTGATCTGCTCGAGCAACACGTACGACAGAAAATGATTGATGCGGCGCAATACAACAAGCAACGCTTCGACAGAGGCAAGGCGAAATTGCGCCCATTTCAACGAGGCGACTACGTACTCGTCAAGAACAATACTCGTAACCAGACATCCCTCGACCTCAAATACAGTGAGCCGTATGAAGTATGCAGAATCCTGGACAACGATCGGTACATCATCAAGCGAGTGACCGGCAGAGGGCGCCCTCGTAAAGTCGCCCACGACCAGTTGCGCCGAGCGCCCCAGCCCGGCGAGCAGGACACCGTGTCGACGGACAGTGTAGAGGACCCGAGCGACAACCGGCGAGCGTGCCGGCCGACGAGACTGAAACAGATGCCACATCTCGGCCCCAGCCCCTGCCGTCTACCTCGCGAGCTGCCCAGGCCTTGGATGCTTAATGGTTAG
Protein
MSEPCDGVGGVSLSDVTFSKNSPEVGAPSSEVGSPPTSARPSGSVNKNMNTSVDGVPADFFTQMLQMMKHVSDRMTSQSDISKVRINDVFLPSYDPDANVGVREWCQHVTIAMETYNLSDYEVRMKAGSLLKGRARLWVDNWLVSTTTWQELRDVLISTFEPENRYSRDVVRFREHIYDNSKDIAQFLSQAWVLWRRITKDKLSNDDAVEAVIGCIGDERLRIELLNARATSVPELISVASSIKSVKRPHPNASIPHQSVAKRPRFDETPGFCQICKRSGHDTRDCRYHDKPEGPQPRQQDGKTTPPGVGRPTCTFCSRLGHTYQTCYKRERAVVSNVNCVGTSKLNAMPVVVGGFKINAIFDSGAECSVIRESVAAKLPGKRVDRVSYLKALGQFTVVSLSTLTTVCIIDNLRVELEFHVVSDIEVSSDILIGINLIENTNLSVVVNSRGAMLVHQPIIYHMRSQSPKFGKLDCDLINDNQISELKILLNKFEHLFIHGYPHTRVNTGELEIRLKDESKTVERRPYRLSPVEREKVRDIVNELLEHKIIRESKSPFASPIIIVKKKNGKDRMCVDYRELNRNTVRDHYPLPIISDQIDQLAGGIYFTTIDMAAGFHQIPISEGSIEKTAFVTPDGLYEYLTMPFGLCNAVSVYQRCINRALAHLTSSPDQVCQVYVDDVLSKCKDFDQGLSHIERILIALQEAGFSINVDKTAFFKRSIEYLGNIIADGQVSPSPRKVEALTKAPIPKTVKQVRQFNGLAGYFRRFIPNFSRVMAPLYELTKNDTTWEWNERHDEARDYIVKHLSTAPTLTLFQEDAPIELYTDASSIGYGAVLVQTFAGRQHPVAYMSQRTTDAESRYHSYELETLAVVRAIKHFRHYLYGRKFKVITDCNALKASKNKKDLLPRIHRWWAFLQNYEFEVEYRKGERLQHADFFSRNPCSEMSVNVMTRDMEWLQIEQHRDDVLRPMMESITSGDSIEGYVMEDGILKKMLTDPILGIHKPIVVPKSFQWSLINSFHVTLQHPGWEKTLQKLRETYWFDKMNSAVRRFVDHCVICRTSKGPSGSIQAQLHPIQKPTAAFQVIHMDITGKLGTRNSEGQYEYVIVAIDAFTKYILLSYSNDKSPSSTLAALKRVIHLFGTPVQVVVDGGREFLGEFKVYTDRIGIDVHSTAPGTSRANGQVERVMGTLKNALTMIKNYSTQDWQTALEALQLAFNCTPHRVTGVAPLTLLTRRQHCIPPELLRLVNIENECIDFDLLEQHVRQKMIDAAQYNKQRFDRGKAKLRPFQRGDYVLVKNNTRNQTSLDLKYSEPYEVCRILDNDRYIIKRVTGRGRPRKVAHDQLRRAPQPGEQDTVSTDSVEDPSDNRRACRPTRLKQMPHLGPSPCRLPRELPRPWMLNG
Summary
Uniprot
B7S8P8
A0A0A9WJJ8
A0A0A9Z954
A0A0A9X1Z8
A0A146M2J2
Q24310
+ More
A0A146KXF1 A0A0A9WTW7 A0A0A9WUU5 A0A0J7KEC9 A0A023EY26 A0A0J7K890 X1X5H2 A0A0J7K9X8 X1WPP0 A0A2S2P032 A0A1Y1MFA9 A0A1W7R6G2 A0A1W7R6L8 J9JJ45 A0A0J7KLK4 A0A023EZG2 A0A023EYH9 A0A0J7NF11 A0A2S2NKJ2 J9LBS9 J9JZ77 T1PMR9 A0A2S2NFQ4 A0A023EYF0 A0A2S2PGQ9 A0A023F013 A0A0A1XB16 X1XU14 A0A224XHR1 A0A034VAM6 A0A034VPR8 J9JQ65 W8APB2 W8C8Q1 W8B0V9 A0A1W7R6H1 A0A2S2P2K5 A0A2M4AEU2 Q24262 A0A0J7KJ08 A0A2S2N9B2
A0A146KXF1 A0A0A9WTW7 A0A0A9WUU5 A0A0J7KEC9 A0A023EY26 A0A0J7K890 X1X5H2 A0A0J7K9X8 X1WPP0 A0A2S2P032 A0A1Y1MFA9 A0A1W7R6G2 A0A1W7R6L8 J9JJ45 A0A0J7KLK4 A0A023EZG2 A0A023EYH9 A0A0J7NF11 A0A2S2NKJ2 J9LBS9 J9JZ77 T1PMR9 A0A2S2NFQ4 A0A023EYF0 A0A2S2PGQ9 A0A023F013 A0A0A1XB16 X1XU14 A0A224XHR1 A0A034VAM6 A0A034VPR8 J9JQ65 W8APB2 W8C8Q1 W8B0V9 A0A1W7R6H1 A0A2S2P2K5 A0A2M4AEU2 Q24262 A0A0J7KJ08 A0A2S2N9B2
EMBL
EF710649
ACE75273.1
GBHO01036023
GBHO01032007
JAG07581.1
JAG11597.1
+ More
GBHO01040010 GBHO01001812 JAG03594.1 JAG41792.1 GBHO01030786 JAG12818.1 GDHC01004980 JAQ13649.1 X14037 CAA32198.1 GDHC01018907 JAP99721.1 GBHO01032415 JAG11189.1 GBHO01032413 JAG11191.1 LBMM01008871 KMQ88569.1 GBBI01004477 JAC14235.1 LBMM01011782 KMQ86633.1 ABLF02004401 ABLF02013544 ABLF02013550 LBMM01010976 KMQ87102.1 ABLF02033943 ABLF02042737 GGMR01009949 MBY22568.1 GEZM01033083 JAV84424.1 GEHC01000888 JAV46757.1 GEHC01000930 JAV46715.1 ABLF02008401 ABLF02008404 LBMM01005919 KMQ91096.1 GBBI01004551 JAC14161.1 GBBI01004550 JAC14162.1 KMQ91095.1 GGMR01005091 MBY17710.1 ABLF02041876 ABLF02014660 ABLF02042774 KA649989 AFP64618.1 GGMR01003388 MBY16007.1 GBBI01004549 JAC14163.1 GGMR01016004 MBY28623.1 GBBI01004613 JAC14099.1 GBXI01005773 JAD08519.1 ABLF02003120 ABLF02004015 ABLF02057179 ABLF02066208 GFTR01008753 JAW07673.1 GAKP01019383 JAC39569.1 GAKP01014483 JAC44469.1 ABLF02005846 GAMC01020207 JAB86348.1 GAMC01007326 JAB99229.1 GAMC01015907 JAB90648.1 GEHC01000876 JAV46769.1 GGMR01011041 MBY23660.1 GGFK01005990 MBW39311.1 Z27119 CAA81643.1 LBMM01006880 KMQ90234.1 GGMR01000717 MBY13336.1
GBHO01040010 GBHO01001812 JAG03594.1 JAG41792.1 GBHO01030786 JAG12818.1 GDHC01004980 JAQ13649.1 X14037 CAA32198.1 GDHC01018907 JAP99721.1 GBHO01032415 JAG11189.1 GBHO01032413 JAG11191.1 LBMM01008871 KMQ88569.1 GBBI01004477 JAC14235.1 LBMM01011782 KMQ86633.1 ABLF02004401 ABLF02013544 ABLF02013550 LBMM01010976 KMQ87102.1 ABLF02033943 ABLF02042737 GGMR01009949 MBY22568.1 GEZM01033083 JAV84424.1 GEHC01000888 JAV46757.1 GEHC01000930 JAV46715.1 ABLF02008401 ABLF02008404 LBMM01005919 KMQ91096.1 GBBI01004551 JAC14161.1 GBBI01004550 JAC14162.1 KMQ91095.1 GGMR01005091 MBY17710.1 ABLF02041876 ABLF02014660 ABLF02042774 KA649989 AFP64618.1 GGMR01003388 MBY16007.1 GBBI01004549 JAC14163.1 GGMR01016004 MBY28623.1 GBBI01004613 JAC14099.1 GBXI01005773 JAD08519.1 ABLF02003120 ABLF02004015 ABLF02057179 ABLF02066208 GFTR01008753 JAW07673.1 GAKP01019383 JAC39569.1 GAKP01014483 JAC44469.1 ABLF02005846 GAMC01020207 JAB86348.1 GAMC01007326 JAB99229.1 GAMC01015907 JAB90648.1 GEHC01000876 JAV46769.1 GGMR01011041 MBY23660.1 GGFK01005990 MBW39311.1 Z27119 CAA81643.1 LBMM01006880 KMQ90234.1 GGMR01000717 MBY13336.1
Proteomes
Pfam
Interpro
IPR041577
RT_RNaseH_2
+ More
IPR001584 Integrase_cat-core
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR041373 RT_RNaseH
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR018061 Retropepsins
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR005162 Retrotrans_gag_dom
IPR019103 Peptidase_aspartic_DDI1-type
IPR028002 Myb_DNA-bind_5
IPR032071 DUF4806
IPR034145 RP_RTVL-H-like
IPR001584 Integrase_cat-core
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR041373 RT_RNaseH
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR018061 Retropepsins
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR005162 Retrotrans_gag_dom
IPR019103 Peptidase_aspartic_DDI1-type
IPR028002 Myb_DNA-bind_5
IPR032071 DUF4806
IPR034145 RP_RTVL-H-like
Gene 3D
ProteinModelPortal
B7S8P8
A0A0A9WJJ8
A0A0A9Z954
A0A0A9X1Z8
A0A146M2J2
Q24310
+ More
A0A146KXF1 A0A0A9WTW7 A0A0A9WUU5 A0A0J7KEC9 A0A023EY26 A0A0J7K890 X1X5H2 A0A0J7K9X8 X1WPP0 A0A2S2P032 A0A1Y1MFA9 A0A1W7R6G2 A0A1W7R6L8 J9JJ45 A0A0J7KLK4 A0A023EZG2 A0A023EYH9 A0A0J7NF11 A0A2S2NKJ2 J9LBS9 J9JZ77 T1PMR9 A0A2S2NFQ4 A0A023EYF0 A0A2S2PGQ9 A0A023F013 A0A0A1XB16 X1XU14 A0A224XHR1 A0A034VAM6 A0A034VPR8 J9JQ65 W8APB2 W8C8Q1 W8B0V9 A0A1W7R6H1 A0A2S2P2K5 A0A2M4AEU2 Q24262 A0A0J7KJ08 A0A2S2N9B2
A0A146KXF1 A0A0A9WTW7 A0A0A9WUU5 A0A0J7KEC9 A0A023EY26 A0A0J7K890 X1X5H2 A0A0J7K9X8 X1WPP0 A0A2S2P032 A0A1Y1MFA9 A0A1W7R6G2 A0A1W7R6L8 J9JJ45 A0A0J7KLK4 A0A023EZG2 A0A023EYH9 A0A0J7NF11 A0A2S2NKJ2 J9LBS9 J9JZ77 T1PMR9 A0A2S2NFQ4 A0A023EYF0 A0A2S2PGQ9 A0A023F013 A0A0A1XB16 X1XU14 A0A224XHR1 A0A034VAM6 A0A034VPR8 J9JQ65 W8APB2 W8C8Q1 W8B0V9 A0A1W7R6H1 A0A2S2P2K5 A0A2M4AEU2 Q24262 A0A0J7KJ08 A0A2S2N9B2
PDB
4OL8
E-value=1.25193e-72,
Score=700
Ontologies
GO
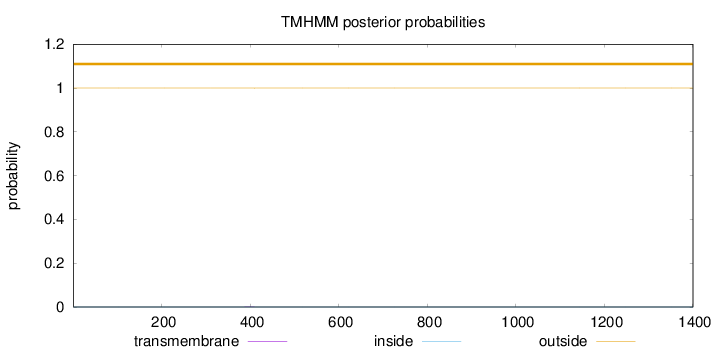
Topology
Length:
1400
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00676999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00026
outside
1 - 1400
Population Genetic Test Statistics
Pi
11.360465
Theta
16.866666
Tajima's D
-1.028499
CLR
39.691183
CSRT
0.133993300334983
Interpretation
Uncertain
