Pre Gene Modal
BGIBMGA012008
Annotation
hypothetical_protein_KGM_10310_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.154 Nuclear Reliability : 1.704
Sequence
CDS
ATGAACTTGATTAAAAGAAGTATTGTTCATTGTAAAAAATGTCTAAAAGGAATTCACAGAAGTTTCAGCCAAAATTGTTGCGTAAAGAATGCGGCAAATGAAAATTCTCAAGTTGAAGAATGGCCACGACGTGTGGTCTCTGGAATCCAACCAACTGGTAGCCTGCATGTTGGGAACTACTTTGGTGCAATCAGACGCTGTGTGCGCTTGCAACAGCAAGGCAATGACTTAACACTGTTTATTGCTGACTTACACTCCTTAACTGCACAGCAGAACCCTGAAGTATTACAAAACAATATCTTGGAACTAACAGCATATTTATTAGCAAGCGGTATTGATCCAGAACAGAGCACATTCTTCGTTCAGTCCGCTGTTACAAGACATGCAGAGCTGTGCTGGCTGCTTGCATGTTTGGCTACTAATGCCAGATTGGCCCATCTACCCCAGTATAAAGAGAAGAGTGCTAAGATGAAGGAAGTGCCCATTGGATTATTGCTTTACCCAGTTTTACAGGCAGCTGACGTGTTGATTTACCGCGGTACCCACGTTCCTGTAGGCGCTGATCAGATACAACACCTGCAGGTTGCCTCGCAGTTGGTTAAGACCTTCCATCATAGATACGGAAAGACATTCCCAACACCGAGACCCCTATTGCCAGACGACGGTAGCGATAGAATACTGAGCCTAAGGGATCCAACAAAGAAAATGTCCAAATCGGATTCTGATCCGAAATCCAGGATCTTGCTGAGCGACACGGATGACGTTATTCACAAGAAAATTCGGAAAGCAGTCACAGATTTTACTCCACAGGTTACATATGATCCAAAAGAACGGCCCGGAGTATCAAATCTTATAAAACTACACTGCCTTGCTGAAGATAAACTCCCTGAAGAAGTAGTTGAAGAATCAGATGGTATGACAACGGCGCAGTATAAAGGTGTTGTTGCAAATGCTCTCTGTGAAGCCCTGCGGCCTGTACGGGAACGGGCAGTCAGTTTGCAAGCCCAGCCAGCCTTCCTGCAAGCTGTGCTGCAGCACGGCGCCGTCCGGGCTCGCCAGCGTGCAGATGAAGTCTATCAGGATGTCACAAACAAACTTGGGTTAGCAGTGCCACGATTACACAGTCTAACCAGAGCCGCCACTCAATGA
Protein
MNLIKRSIVHCKKCLKGIHRSFSQNCCVKNAANENSQVEEWPRRVVSGIQPTGSLHVGNYFGAIRRCVRLQQQGNDLTLFIADLHSLTAQQNPEVLQNNILELTAYLLASGIDPEQSTFFVQSAVTRHAELCWLLACLATNARLAHLPQYKEKSAKMKEVPIGLLLYPVLQAADVLIYRGTHVPVGADQIQHLQVASQLVKTFHHRYGKTFPTPRPLLPDDGSDRILSLRDPTKKMSKSDSDPKSRILLSDTDDVIHKKIRKAVTDFTPQVTYDPKERPGVSNLIKLHCLAEDKLPEEVVEESDGMTTAQYKGVVANALCEALRPVRERAVSLQAQPAFLQAVLQHGAVRARQRADEVYQDVTNKLGLAVPRLHSLTRAATQ
Summary
Similarity
Belongs to the class-I aminoacyl-tRNA synthetase family.
Uniprot
H9JR48
A0A212FLW9
A0A3S2TP45
A0A2W1BJG4
A0A194QPI1
A0A2H1WGB6
+ More
B0X7D9 A0A1Q3F732 Q174W2 A0A1B0EX65 A0A1S4FEI3 A0A182NPR3 A0A182MGI9 A0A182K749 A0A1B6DU64 A0A2M4BRG0 A0A2M4AV48 A0A2M4BRB9 A0A1B0CS17 A0A182G0A6 A0A1L8DFR2 A0A2M4BSK5 A0A182YL32 A0A084VIM0 A0A182GWZ9 A0A2C9GQD2 A0A182Q0U9 A0A182VCG0 A0A182TKQ1 A0A182L4L7 A0A1W4X5F1 T1DRF9 A0A232FL39 A0A067RAV5 K7J475 A0A224XSN4 A0A0L0CK73 A0A0L7R0D3 E9J035 A0A182WEB7 A0A023F885 T1DQQ8 T1HZQ2 A0A0P4VQN6 R4G5Y4 T1JAT1 A0A158NB62 A0A182S085 W5JCS0 A0A0V0GAG3 A0A182P6W8 A0A182SYJ5 J9JPX7 E2ARE7 A0A026WQJ1 E9GVX8 A0A1J1IIC9 D6WMA7 A0A182X6Q0 A0A1I8N612 J3JY07 A0A1W4W4C0 Q8SZU2 A0A0M4EZM7 B4J059 B3M3U8 A0A0P5CHY6 T1PJ78 A0A164KFU3 A0A0P6DKE0 B3NDC2 A0A1W4X5B3 A0A2A3EQU5 A0A0P5D3H2 A0A336LWJ8 B4PJH8 A0A0P4YUY2 A0A0K8UDE4 A0A034VKR2 Q9VVL8 B4N4T4 A0A088ANW0 A0A1B6CYZ5 A0A0P6BZ56 W5MYQ0 B4QPB8 Q3YMW3 B4LIF1 A0A0P5FGA9 A0A3B0J4N7 A0A0A1WG87 B4HLB5 Q2LZ15 B4GZR8 A0A195BSS7 C3YIC1 A0A1Y1LB20 A0A195D6H6 W5MYQ9
B0X7D9 A0A1Q3F732 Q174W2 A0A1B0EX65 A0A1S4FEI3 A0A182NPR3 A0A182MGI9 A0A182K749 A0A1B6DU64 A0A2M4BRG0 A0A2M4AV48 A0A2M4BRB9 A0A1B0CS17 A0A182G0A6 A0A1L8DFR2 A0A2M4BSK5 A0A182YL32 A0A084VIM0 A0A182GWZ9 A0A2C9GQD2 A0A182Q0U9 A0A182VCG0 A0A182TKQ1 A0A182L4L7 A0A1W4X5F1 T1DRF9 A0A232FL39 A0A067RAV5 K7J475 A0A224XSN4 A0A0L0CK73 A0A0L7R0D3 E9J035 A0A182WEB7 A0A023F885 T1DQQ8 T1HZQ2 A0A0P4VQN6 R4G5Y4 T1JAT1 A0A158NB62 A0A182S085 W5JCS0 A0A0V0GAG3 A0A182P6W8 A0A182SYJ5 J9JPX7 E2ARE7 A0A026WQJ1 E9GVX8 A0A1J1IIC9 D6WMA7 A0A182X6Q0 A0A1I8N612 J3JY07 A0A1W4W4C0 Q8SZU2 A0A0M4EZM7 B4J059 B3M3U8 A0A0P5CHY6 T1PJ78 A0A164KFU3 A0A0P6DKE0 B3NDC2 A0A1W4X5B3 A0A2A3EQU5 A0A0P5D3H2 A0A336LWJ8 B4PJH8 A0A0P4YUY2 A0A0K8UDE4 A0A034VKR2 Q9VVL8 B4N4T4 A0A088ANW0 A0A1B6CYZ5 A0A0P6BZ56 W5MYQ0 B4QPB8 Q3YMW3 B4LIF1 A0A0P5FGA9 A0A3B0J4N7 A0A0A1WG87 B4HLB5 Q2LZ15 B4GZR8 A0A195BSS7 C3YIC1 A0A1Y1LB20 A0A195D6H6 W5MYQ9
Pubmed
19121390
22118469
28756777
26354079
17510324
25244985
+ More
24438588 26483478 20966253 28648823 24845553 20075255 26108605 21282665 25474469 27129103 21347285 20920257 23761445 20798317 24508170 30249741 21292972 18362917 19820115 25315136 22516182 17994087 17550304 25348373 10731132 12537568 12537572 12537573 12537574 15917496 16110336 17569856 17569867 26109357 26109356 22936249 25830018 15632085 18563158 28004739
24438588 26483478 20966253 28648823 24845553 20075255 26108605 21282665 25474469 27129103 21347285 20920257 23761445 20798317 24508170 30249741 21292972 18362917 19820115 25315136 22516182 17994087 17550304 25348373 10731132 12537568 12537572 12537573 12537574 15917496 16110336 17569856 17569867 26109357 26109356 22936249 25830018 15632085 18563158 28004739
EMBL
BABH01039741
AGBW02007695
OWR54732.1
RSAL01000034
RVE51363.1
KZ150147
+ More
PZC72880.1 KQ461185 KPJ07412.1 ODYU01008482 SOQ52118.1 DS232448 EDS41889.1 GFDL01011747 JAV23298.1 CH477405 EAT41602.1 AJVK01031329 AXCM01013263 GEDC01008095 JAS29203.1 GGFJ01006422 MBW55563.1 GGFK01011333 MBW44654.1 GGFJ01006421 MBW55562.1 AJWK01025563 GFDF01008867 JAV05217.1 GGFJ01006832 MBW55973.1 ATLV01013369 KE524854 KFB37814.1 JXUM01094416 KQ564127 KXJ72729.1 APCN01002389 AXCN02001663 GAMD01001944 JAA99646.1 NNAY01000080 OXU31168.1 KK852584 KDR20847.1 AAZX01003267 GFTR01005293 JAW11133.1 JRES01000296 KNC32656.1 KQ414670 KOC64324.1 GL767291 EFZ13831.1 GBBI01001230 JAC17482.1 GAMD01001020 JAB00571.1 ACPB03021503 GDKW01002439 JAI54156.1 GAHY01000115 JAA77395.1 JH432004 ADTU01010967 ADMH02001767 ETN61138.1 GECL01001073 JAP05051.1 ABLF02035743 GL442063 EFN63956.1 KK107128 QOIP01000006 EZA58290.1 RLU21623.1 GL732569 EFX76370.1 CVRI01000048 CRK98822.1 KQ971343 EFA03333.2 BT128130 AEE63091.1 AY070507 AAL47978.1 CP012525 ALC43870.1 CH916366 EDV97852.1 CH902618 EDV39282.1 GDIP01185943 JAJ37459.1 KA648857 AFP63486.1 LRGB01003325 KZS03226.1 GDIQ01077554 JAN17183.1 CH954178 EDV51915.1 KZ288193 PBC34060.1 GDIP01163343 JAJ60059.1 UFQT01000188 SSX21351.1 CM000159 EDW94666.1 GDIP01223284 JAJ00118.1 GDHF01027929 JAI24385.1 GAKP01015056 JAC43896.1 AE014296 DQ062762 BT044176 AAF49293.1 AAY56635.1 ACH92241.1 AGB94694.1 CH964095 EDW79158.1 GEDC01018631 JAS18667.1 GDIP01007347 JAM96368.1 AHAT01003663 AHAT01003664 CM000363 CM002912 EDX10912.1 KMZ00326.1 DQ062763 AAY56636.1 CH940647 EDW70738.2 GDIQ01255718 JAJ96006.1 OUUW01000002 SPP76495.1 GBXI01016879 JAC97412.1 CH480815 EDW41935.1 CH379069 EAL29694.3 CH479199 EDW29495.1 KQ976417 KYM89723.1 GG666514 EEN60258.1 GEZM01060842 JAV70859.1 KQ976760 KYN08500.1
PZC72880.1 KQ461185 KPJ07412.1 ODYU01008482 SOQ52118.1 DS232448 EDS41889.1 GFDL01011747 JAV23298.1 CH477405 EAT41602.1 AJVK01031329 AXCM01013263 GEDC01008095 JAS29203.1 GGFJ01006422 MBW55563.1 GGFK01011333 MBW44654.1 GGFJ01006421 MBW55562.1 AJWK01025563 GFDF01008867 JAV05217.1 GGFJ01006832 MBW55973.1 ATLV01013369 KE524854 KFB37814.1 JXUM01094416 KQ564127 KXJ72729.1 APCN01002389 AXCN02001663 GAMD01001944 JAA99646.1 NNAY01000080 OXU31168.1 KK852584 KDR20847.1 AAZX01003267 GFTR01005293 JAW11133.1 JRES01000296 KNC32656.1 KQ414670 KOC64324.1 GL767291 EFZ13831.1 GBBI01001230 JAC17482.1 GAMD01001020 JAB00571.1 ACPB03021503 GDKW01002439 JAI54156.1 GAHY01000115 JAA77395.1 JH432004 ADTU01010967 ADMH02001767 ETN61138.1 GECL01001073 JAP05051.1 ABLF02035743 GL442063 EFN63956.1 KK107128 QOIP01000006 EZA58290.1 RLU21623.1 GL732569 EFX76370.1 CVRI01000048 CRK98822.1 KQ971343 EFA03333.2 BT128130 AEE63091.1 AY070507 AAL47978.1 CP012525 ALC43870.1 CH916366 EDV97852.1 CH902618 EDV39282.1 GDIP01185943 JAJ37459.1 KA648857 AFP63486.1 LRGB01003325 KZS03226.1 GDIQ01077554 JAN17183.1 CH954178 EDV51915.1 KZ288193 PBC34060.1 GDIP01163343 JAJ60059.1 UFQT01000188 SSX21351.1 CM000159 EDW94666.1 GDIP01223284 JAJ00118.1 GDHF01027929 JAI24385.1 GAKP01015056 JAC43896.1 AE014296 DQ062762 BT044176 AAF49293.1 AAY56635.1 ACH92241.1 AGB94694.1 CH964095 EDW79158.1 GEDC01018631 JAS18667.1 GDIP01007347 JAM96368.1 AHAT01003663 AHAT01003664 CM000363 CM002912 EDX10912.1 KMZ00326.1 DQ062763 AAY56636.1 CH940647 EDW70738.2 GDIQ01255718 JAJ96006.1 OUUW01000002 SPP76495.1 GBXI01016879 JAC97412.1 CH480815 EDW41935.1 CH379069 EAL29694.3 CH479199 EDW29495.1 KQ976417 KYM89723.1 GG666514 EEN60258.1 GEZM01060842 JAV70859.1 KQ976760 KYN08500.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000053240
UP000002320
UP000008820
+ More
UP000092462 UP000075884 UP000075883 UP000075881 UP000092461 UP000069272 UP000076408 UP000030765 UP000069940 UP000249989 UP000075840 UP000075886 UP000075903 UP000075902 UP000075882 UP000192223 UP000215335 UP000027135 UP000002358 UP000037069 UP000053825 UP000075920 UP000015103 UP000005205 UP000075900 UP000000673 UP000075885 UP000075901 UP000007819 UP000000311 UP000053097 UP000279307 UP000000305 UP000183832 UP000007266 UP000076407 UP000095301 UP000192221 UP000092553 UP000001070 UP000007801 UP000076858 UP000008711 UP000242457 UP000002282 UP000000803 UP000007798 UP000005203 UP000018468 UP000000304 UP000008792 UP000268350 UP000001292 UP000001819 UP000008744 UP000078540 UP000001554 UP000078542
UP000092462 UP000075884 UP000075883 UP000075881 UP000092461 UP000069272 UP000076408 UP000030765 UP000069940 UP000249989 UP000075840 UP000075886 UP000075903 UP000075902 UP000075882 UP000192223 UP000215335 UP000027135 UP000002358 UP000037069 UP000053825 UP000075920 UP000015103 UP000005205 UP000075900 UP000000673 UP000075885 UP000075901 UP000007819 UP000000311 UP000053097 UP000279307 UP000000305 UP000183832 UP000007266 UP000076407 UP000095301 UP000192221 UP000092553 UP000001070 UP000007801 UP000076858 UP000008711 UP000242457 UP000002282 UP000000803 UP000007798 UP000005203 UP000018468 UP000000304 UP000008792 UP000268350 UP000001292 UP000001819 UP000008744 UP000078540 UP000001554 UP000078542
Interpro
IPR014729
Rossmann-like_a/b/a_fold
+ More
IPR002305 aa-tRNA-synth_Ic
IPR001412 aa-tRNA-synth_I_CS
IPR024109 Trp-tRNA-ligase_bac-type
IPR002306 Trp-tRNA-ligase
IPR003593 AAA+_ATPase
IPR000048 IQ_motif_EF-hand-BS
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR000742 EGF-like_dom
IPR013032 EGF-like_CS
IPR038677 WIF_sf
IPR003306 WIF
IPR002305 aa-tRNA-synth_Ic
IPR001412 aa-tRNA-synth_I_CS
IPR024109 Trp-tRNA-ligase_bac-type
IPR002306 Trp-tRNA-ligase
IPR003593 AAA+_ATPase
IPR000048 IQ_motif_EF-hand-BS
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR000742 EGF-like_dom
IPR013032 EGF-like_CS
IPR038677 WIF_sf
IPR003306 WIF
Gene 3D
ProteinModelPortal
H9JR48
A0A212FLW9
A0A3S2TP45
A0A2W1BJG4
A0A194QPI1
A0A2H1WGB6
+ More
B0X7D9 A0A1Q3F732 Q174W2 A0A1B0EX65 A0A1S4FEI3 A0A182NPR3 A0A182MGI9 A0A182K749 A0A1B6DU64 A0A2M4BRG0 A0A2M4AV48 A0A2M4BRB9 A0A1B0CS17 A0A182G0A6 A0A1L8DFR2 A0A2M4BSK5 A0A182YL32 A0A084VIM0 A0A182GWZ9 A0A2C9GQD2 A0A182Q0U9 A0A182VCG0 A0A182TKQ1 A0A182L4L7 A0A1W4X5F1 T1DRF9 A0A232FL39 A0A067RAV5 K7J475 A0A224XSN4 A0A0L0CK73 A0A0L7R0D3 E9J035 A0A182WEB7 A0A023F885 T1DQQ8 T1HZQ2 A0A0P4VQN6 R4G5Y4 T1JAT1 A0A158NB62 A0A182S085 W5JCS0 A0A0V0GAG3 A0A182P6W8 A0A182SYJ5 J9JPX7 E2ARE7 A0A026WQJ1 E9GVX8 A0A1J1IIC9 D6WMA7 A0A182X6Q0 A0A1I8N612 J3JY07 A0A1W4W4C0 Q8SZU2 A0A0M4EZM7 B4J059 B3M3U8 A0A0P5CHY6 T1PJ78 A0A164KFU3 A0A0P6DKE0 B3NDC2 A0A1W4X5B3 A0A2A3EQU5 A0A0P5D3H2 A0A336LWJ8 B4PJH8 A0A0P4YUY2 A0A0K8UDE4 A0A034VKR2 Q9VVL8 B4N4T4 A0A088ANW0 A0A1B6CYZ5 A0A0P6BZ56 W5MYQ0 B4QPB8 Q3YMW3 B4LIF1 A0A0P5FGA9 A0A3B0J4N7 A0A0A1WG87 B4HLB5 Q2LZ15 B4GZR8 A0A195BSS7 C3YIC1 A0A1Y1LB20 A0A195D6H6 W5MYQ9
B0X7D9 A0A1Q3F732 Q174W2 A0A1B0EX65 A0A1S4FEI3 A0A182NPR3 A0A182MGI9 A0A182K749 A0A1B6DU64 A0A2M4BRG0 A0A2M4AV48 A0A2M4BRB9 A0A1B0CS17 A0A182G0A6 A0A1L8DFR2 A0A2M4BSK5 A0A182YL32 A0A084VIM0 A0A182GWZ9 A0A2C9GQD2 A0A182Q0U9 A0A182VCG0 A0A182TKQ1 A0A182L4L7 A0A1W4X5F1 T1DRF9 A0A232FL39 A0A067RAV5 K7J475 A0A224XSN4 A0A0L0CK73 A0A0L7R0D3 E9J035 A0A182WEB7 A0A023F885 T1DQQ8 T1HZQ2 A0A0P4VQN6 R4G5Y4 T1JAT1 A0A158NB62 A0A182S085 W5JCS0 A0A0V0GAG3 A0A182P6W8 A0A182SYJ5 J9JPX7 E2ARE7 A0A026WQJ1 E9GVX8 A0A1J1IIC9 D6WMA7 A0A182X6Q0 A0A1I8N612 J3JY07 A0A1W4W4C0 Q8SZU2 A0A0M4EZM7 B4J059 B3M3U8 A0A0P5CHY6 T1PJ78 A0A164KFU3 A0A0P6DKE0 B3NDC2 A0A1W4X5B3 A0A2A3EQU5 A0A0P5D3H2 A0A336LWJ8 B4PJH8 A0A0P4YUY2 A0A0K8UDE4 A0A034VKR2 Q9VVL8 B4N4T4 A0A088ANW0 A0A1B6CYZ5 A0A0P6BZ56 W5MYQ0 B4QPB8 Q3YMW3 B4LIF1 A0A0P5FGA9 A0A3B0J4N7 A0A0A1WG87 B4HLB5 Q2LZ15 B4GZR8 A0A195BSS7 C3YIC1 A0A1Y1LB20 A0A195D6H6 W5MYQ9
PDB
5EKD
E-value=3.41171e-73,
Score=699
Ontologies
GO
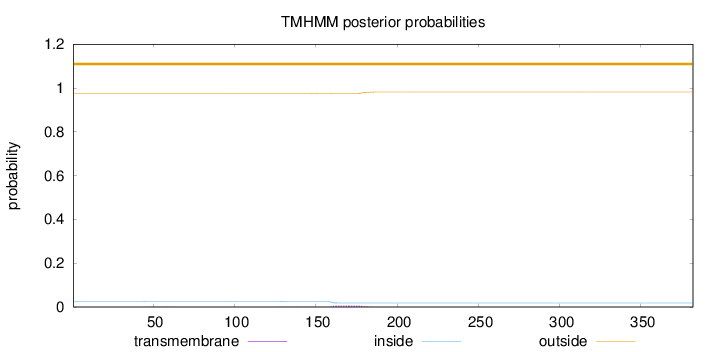
Topology
Length:
382
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13872
Exp number, first 60 AAs:
0.0021
Total prob of N-in:
0.02484
outside
1 - 382
Population Genetic Test Statistics
Pi
24.531539
Theta
22.109374
Tajima's D
0.471247
CLR
0.905563
CSRT
0.510824458777061
Interpretation
Uncertain
