Gene
KWMTBOMO06893 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012007
Annotation
TAR_DNA_binding_protein_homolog_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.11 PlasmaMembrane Reliability : 1.163
Sequence
CDS
ATGACGGAGTACGTGCCGGTAAGCGAGGGCGAGCAAGACGAGCCCATTGAGCTGCCAACCGAAGAAGACGGCTCGCTGCTACTGTCGACCGTGGCAGCGCAGTTCGCAGGTGCCAGCGGCCTGAAGTACCGAGTGAACGGGAGACTACGCGGCGTACGCCTCGTCGACGAGCGCCTGTCACCACCGGCGGAGGGCTGGCCGGCCTACCGATATTGGTGTGCCTTCCCACGCGCGGAATCGCCGCCGCCTCGACCACGATGCTCCGATCTTATAGTTCTCGGCTTACCATGGAAGACGACAGAAGAAGCTGTACGAGATTACTTCTCCTCCTTTGGCGATTTGCTGATGGTTCAAGTAAAACGCGATACGCGCACCGGACTATCCAAAGGTTTTGGCTTCATCAAATTCGCCGAGTACGAAGCTCAACTGCGTGCATTAGGCCGCCGACACATGATAGACGGGCGCTGGTGCGATGTTCGCATCCCGAACGGCAAGGACGGCTCGTCGACGGCTCATCGAAAGGTATTCGTCGGCCGTTGTACGGAGAATATTACAGCCGAAGATCTCAGAGAGTACTTCGGCTCCTTTGGTCAAGTGACAGATGTGTTTGTGCCGCGACCCTTTCGTGCATTCAGCTTTGTGACGTTTCTTGACCCAGAAGTAGCAAGGTCCTTGTGTGGACAAGATCATATTATAAAAGGTGTGTCAGTAAACATTTCAACAGCATCACCAAAGCGCGAAAGGTCCACTACTGCAGCCAGTGGGCTCGGCTGGGGCCGTATGCTTGGCTGGCCAGCTGATGCATGGCCACTCCCGGCACCCCATCCCCTCGAGTCAAAGCCCTACCTCAAATACGAGTAA
Protein
MTEYVPVSEGEQDEPIELPTEEDGSLLLSTVAAQFAGASGLKYRVNGRLRGVRLVDERLSPPAEGWPAYRYWCAFPRAESPPPRPRCSDLIVLGLPWKTTEEAVRDYFSSFGDLLMVQVKRDTRTGLSKGFGFIKFAEYEAQLRALGRRHMIDGRWCDVRIPNGKDGSSTAHRKVFVGRCTENITAEDLREYFGSFGQVTDVFVPRPFRAFSFVTFLDPEVARSLCGQDHIIKGVSVNISTASPKRERSTTAASGLGWGRMLGWPADAWPLPAPHPLESKPYLKYE
Summary
Uniprot
H9JR47
Q1HDZ2
A0A2H1WGD2
A0A2W1BC91
A0A2A4JD38
S4P175
+ More
A0A194QV33 A0A212FM15 A0A194Q9L2 A0A0L7KN35 A0A2H1VAS8 A0A0L7KS51 A0A0L8HEQ9 A0A0T6BDA3 A0A1Y1N911 A0A2T7PUB6 V9ICK0 A0A1Y1NDZ0 A0A0B7A1I3 A0A0Q9WQA6 D6WYQ8 Q9W1I0 A0A1Y1NCY2 R7TFC3 T1J0V8 A0A194QKQ3 O97469 B4I8U6 B4QB08 A0A1I8PU82 A0A1B6F1X4 B4PA81 E0W1D8 A0A1B6FIW6 A0A1D2MYP4 Q8SXP8 A0A1W4V1I0 B3NPT7 H9JBD7 A0A1B6EET0 A0A1I8PU56 A0A1E1W3L5 A0A1B6DH86 O97468 A0A0R3P1I8 A0A3B0JKJ8 A0A1B6J4B6 A0A1B6FA14 V5GJJ3 A0A0P5ETW0 K7J2F6 B4MRQ1 A0A1B6MTC6 A0A0P5GIW9 A0A1W4UZX8 A0A212EIJ4 A0A1B6IJ03 A0A1B6DFI4 A0A1W4UMZ9 A0A0A9Z220 A0A0P5H4E6 V5HVY6 A0A0Q9W423 A0A194PK77 A0A0P5G601 A0A2T7PSN6 A0A336M2M6 A0A131ZA78 A0A0P6E442 B3MEM3 A0A023GB93 A0A336MQ24 B4KMD9 B4J632 A0A3B0JD50 B4GCQ9 Q29CQ4 A0A0P5JNR3 T1HYV6 A0A067RFQ3 A0A0Q9W4P3 A0A1B6CB61 A0A224XBX8 A0A069DV00 A0A2J7QU24 A0A0P5GU05 A0A210PJN0 V5GVK5 A0A0K8TS76 A0A0P6DBT1 B4LNX4 A0A3S0ZIL9 A0A195EHV4 F4W8F4 A0A195FB69 A0A158NK58 A0A151X6Y2 A0A0P5GQY4 A0A0B7A069 A0A131Y1K6
A0A194QV33 A0A212FM15 A0A194Q9L2 A0A0L7KN35 A0A2H1VAS8 A0A0L7KS51 A0A0L8HEQ9 A0A0T6BDA3 A0A1Y1N911 A0A2T7PUB6 V9ICK0 A0A1Y1NDZ0 A0A0B7A1I3 A0A0Q9WQA6 D6WYQ8 Q9W1I0 A0A1Y1NCY2 R7TFC3 T1J0V8 A0A194QKQ3 O97469 B4I8U6 B4QB08 A0A1I8PU82 A0A1B6F1X4 B4PA81 E0W1D8 A0A1B6FIW6 A0A1D2MYP4 Q8SXP8 A0A1W4V1I0 B3NPT7 H9JBD7 A0A1B6EET0 A0A1I8PU56 A0A1E1W3L5 A0A1B6DH86 O97468 A0A0R3P1I8 A0A3B0JKJ8 A0A1B6J4B6 A0A1B6FA14 V5GJJ3 A0A0P5ETW0 K7J2F6 B4MRQ1 A0A1B6MTC6 A0A0P5GIW9 A0A1W4UZX8 A0A212EIJ4 A0A1B6IJ03 A0A1B6DFI4 A0A1W4UMZ9 A0A0A9Z220 A0A0P5H4E6 V5HVY6 A0A0Q9W423 A0A194PK77 A0A0P5G601 A0A2T7PSN6 A0A336M2M6 A0A131ZA78 A0A0P6E442 B3MEM3 A0A023GB93 A0A336MQ24 B4KMD9 B4J632 A0A3B0JD50 B4GCQ9 Q29CQ4 A0A0P5JNR3 T1HYV6 A0A067RFQ3 A0A0Q9W4P3 A0A1B6CB61 A0A224XBX8 A0A069DV00 A0A2J7QU24 A0A0P5GU05 A0A210PJN0 V5GVK5 A0A0K8TS76 A0A0P6DBT1 B4LNX4 A0A3S0ZIL9 A0A195EHV4 F4W8F4 A0A195FB69 A0A158NK58 A0A151X6Y2 A0A0P5GQY4 A0A0B7A069 A0A131Y1K6
Pubmed
19121390
30836863
28756777
23622113
26354079
22118469
+ More
26227816 28004739 17994087 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23254933 10191082 22936249 17550304 20566863 27289101 26109357 26109356 18057021 15632085 20075255 25401762 26823975 25765539 26830274 23185243 24845553 26334808 28812685 26369729 21719571 21347285
26227816 28004739 17994087 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23254933 10191082 22936249 17550304 20566863 27289101 26109357 26109356 18057021 15632085 20075255 25401762 26823975 25765539 26830274 23185243 24845553 26334808 28812685 26369729 21719571 21347285
EMBL
BABH01039741
BABH01039742
BABH01039743
BABH01039744
DQ497205
MH745576
+ More
ABF55970.1 QBM91283.1 ODYU01008482 SOQ52121.1 KZ150147 PZC72882.1 NWSH01002002 PCG69464.1 GAIX01009031 JAA83529.1 KQ461185 KPJ07416.1 AGBW02007695 OWR54729.1 KQ459252 KPJ02202.1 JTDY01008189 KOB64688.1 ODYU01001561 SOQ37948.1 JTDY01006591 KOB65854.1 KQ418333 KOF87728.1 LJIG01001651 KRT85289.1 GEZM01009643 JAV94434.1 PZQS01000002 PVD37008.1 JR038232 AEY58171.1 GEZM01009641 JAV94436.1 HACG01027076 CEK73941.1 CH963850 KRF98137.1 KQ971357 EFA08408.2 AE013599 AAF47080.2 GEZM01009642 JAV94435.1 AMQN01014735 KB311222 ELT89721.1 AFFK01020674 KQ461198 KPJ06102.1 AB010261 AB019707 BAA34430.1 CH480824 EDW57021.1 CM000362 CM002911 EDX08445.1 KMY96135.1 GECZ01025510 JAS44259.1 CM000158 EDW92407.1 KRK00493.1 KRK00494.1 KRK00495.1 KRK00496.1 DS235870 EEB19444.1 GECZ01019627 JAS50142.1 LJIJ01000375 ODM98169.1 AY089497 BT022083 AAF47078.2 AAF47079.2 AAL90235.1 AAM68274.1 AAY33499.1 ACZ94552.1 ACZ94553.1 CH954179 EDV56878.1 KQS63062.1 KQS63063.1 KQS63064.1 KQS63065.1 BABH01030827 GEDC01000870 JAS36428.1 GDQN01009477 JAT81577.1 GEDC01012251 JAS25047.1 AB019705 AB019706 BAA34429.1 CH475433 KRT05136.1 OUUW01000001 SPP73151.1 GECU01013681 GECU01001932 JAS94025.1 JAT05775.1 GECZ01022742 JAS47027.1 GALX01004197 JAB64269.1 GDIQ01268046 JAJ83678.1 EDW74790.1 GEBQ01000785 JAT39192.1 GDIQ01240888 JAK10837.1 AGBW02014631 OWR41291.1 GECU01020815 JAS86891.1 GEDC01012861 JAS24437.1 GBHO01007744 GBRD01016884 GDHC01004483 JAG35860.1 JAG48943.1 JAQ14146.1 GDIQ01239198 JAK12527.1 GANP01005900 JAB78568.1 CH940648 KRF79811.1 KQ459606 KPI91500.1 GDIQ01248928 JAK02797.1 PVD36397.1 UFQT01000450 SSX24495.1 GEDV01000314 JAP88243.1 GDIQ01082847 JAN11890.1 CH902619 EDV35487.1 KPU75688.1 KPU75689.1 GBBM01003962 JAC31456.1 UFQT01001404 SSX30397.1 CH933808 EDW09827.1 KRG04921.1 CH916367 EDW01890.1 SPP73150.1 CH479181 EDW31507.1 EAL29286.2 KRT05137.1 KRT05138.1 KRT05139.1 KRT05140.1 GDIQ01222142 JAK29583.1 ACPB03009442 KK852531 KDR21878.1 KRF79809.1 KRF79810.1 KRF79812.1 KRF79813.1 GEDC01026808 JAS10490.1 GFTR01006510 JAW09916.1 GBGD01001362 JAC87527.1 NEVH01011190 PNF32092.1 GDIQ01242648 JAK09077.1 NEDP02076495 OWF36684.1 GALX01004198 JAB64268.1 GDAI01000818 JAI16785.1 GDIQ01081294 JAN13443.1 EDW61143.1 RQTK01000497 RUS78651.1 KQ978881 KYN27833.1 GL887908 EGI69445.1 KQ981693 KYN37626.1 ADTU01018711 ADTU01018712 ADTU01018713 ADTU01018714 KQ982472 KYQ56079.1 GDIQ01243892 JAK07833.1 HACG01027077 CEK73942.1 GEFM01002423 JAP73373.1
ABF55970.1 QBM91283.1 ODYU01008482 SOQ52121.1 KZ150147 PZC72882.1 NWSH01002002 PCG69464.1 GAIX01009031 JAA83529.1 KQ461185 KPJ07416.1 AGBW02007695 OWR54729.1 KQ459252 KPJ02202.1 JTDY01008189 KOB64688.1 ODYU01001561 SOQ37948.1 JTDY01006591 KOB65854.1 KQ418333 KOF87728.1 LJIG01001651 KRT85289.1 GEZM01009643 JAV94434.1 PZQS01000002 PVD37008.1 JR038232 AEY58171.1 GEZM01009641 JAV94436.1 HACG01027076 CEK73941.1 CH963850 KRF98137.1 KQ971357 EFA08408.2 AE013599 AAF47080.2 GEZM01009642 JAV94435.1 AMQN01014735 KB311222 ELT89721.1 AFFK01020674 KQ461198 KPJ06102.1 AB010261 AB019707 BAA34430.1 CH480824 EDW57021.1 CM000362 CM002911 EDX08445.1 KMY96135.1 GECZ01025510 JAS44259.1 CM000158 EDW92407.1 KRK00493.1 KRK00494.1 KRK00495.1 KRK00496.1 DS235870 EEB19444.1 GECZ01019627 JAS50142.1 LJIJ01000375 ODM98169.1 AY089497 BT022083 AAF47078.2 AAF47079.2 AAL90235.1 AAM68274.1 AAY33499.1 ACZ94552.1 ACZ94553.1 CH954179 EDV56878.1 KQS63062.1 KQS63063.1 KQS63064.1 KQS63065.1 BABH01030827 GEDC01000870 JAS36428.1 GDQN01009477 JAT81577.1 GEDC01012251 JAS25047.1 AB019705 AB019706 BAA34429.1 CH475433 KRT05136.1 OUUW01000001 SPP73151.1 GECU01013681 GECU01001932 JAS94025.1 JAT05775.1 GECZ01022742 JAS47027.1 GALX01004197 JAB64269.1 GDIQ01268046 JAJ83678.1 EDW74790.1 GEBQ01000785 JAT39192.1 GDIQ01240888 JAK10837.1 AGBW02014631 OWR41291.1 GECU01020815 JAS86891.1 GEDC01012861 JAS24437.1 GBHO01007744 GBRD01016884 GDHC01004483 JAG35860.1 JAG48943.1 JAQ14146.1 GDIQ01239198 JAK12527.1 GANP01005900 JAB78568.1 CH940648 KRF79811.1 KQ459606 KPI91500.1 GDIQ01248928 JAK02797.1 PVD36397.1 UFQT01000450 SSX24495.1 GEDV01000314 JAP88243.1 GDIQ01082847 JAN11890.1 CH902619 EDV35487.1 KPU75688.1 KPU75689.1 GBBM01003962 JAC31456.1 UFQT01001404 SSX30397.1 CH933808 EDW09827.1 KRG04921.1 CH916367 EDW01890.1 SPP73150.1 CH479181 EDW31507.1 EAL29286.2 KRT05137.1 KRT05138.1 KRT05139.1 KRT05140.1 GDIQ01222142 JAK29583.1 ACPB03009442 KK852531 KDR21878.1 KRF79809.1 KRF79810.1 KRF79812.1 KRF79813.1 GEDC01026808 JAS10490.1 GFTR01006510 JAW09916.1 GBGD01001362 JAC87527.1 NEVH01011190 PNF32092.1 GDIQ01242648 JAK09077.1 NEDP02076495 OWF36684.1 GALX01004198 JAB64268.1 GDAI01000818 JAI16785.1 GDIQ01081294 JAN13443.1 EDW61143.1 RQTK01000497 RUS78651.1 KQ978881 KYN27833.1 GL887908 EGI69445.1 KQ981693 KYN37626.1 ADTU01018711 ADTU01018712 ADTU01018713 ADTU01018714 KQ982472 KYQ56079.1 GDIQ01243892 JAK07833.1 HACG01027077 CEK73942.1 GEFM01002423 JAP73373.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000053454 UP000245119 UP000007798 UP000007266 UP000000803 UP000014760 UP000001292 UP000000304 UP000095300 UP000002282 UP000009046 UP000094527 UP000192221 UP000008711 UP000001819 UP000268350 UP000002358 UP000008792 UP000007801 UP000009192 UP000001070 UP000008744 UP000015103 UP000027135 UP000235965 UP000242188 UP000271974 UP000078492 UP000007755 UP000078541 UP000005205 UP000075809
UP000053454 UP000245119 UP000007798 UP000007266 UP000000803 UP000014760 UP000001292 UP000000304 UP000095300 UP000002282 UP000009046 UP000094527 UP000192221 UP000008711 UP000001819 UP000268350 UP000002358 UP000008792 UP000007801 UP000009192 UP000001070 UP000008744 UP000015103 UP000027135 UP000235965 UP000242188 UP000271974 UP000078492 UP000007755 UP000078541 UP000005205 UP000075809
Interpro
Gene 3D
ProteinModelPortal
H9JR47
Q1HDZ2
A0A2H1WGD2
A0A2W1BC91
A0A2A4JD38
S4P175
+ More
A0A194QV33 A0A212FM15 A0A194Q9L2 A0A0L7KN35 A0A2H1VAS8 A0A0L7KS51 A0A0L8HEQ9 A0A0T6BDA3 A0A1Y1N911 A0A2T7PUB6 V9ICK0 A0A1Y1NDZ0 A0A0B7A1I3 A0A0Q9WQA6 D6WYQ8 Q9W1I0 A0A1Y1NCY2 R7TFC3 T1J0V8 A0A194QKQ3 O97469 B4I8U6 B4QB08 A0A1I8PU82 A0A1B6F1X4 B4PA81 E0W1D8 A0A1B6FIW6 A0A1D2MYP4 Q8SXP8 A0A1W4V1I0 B3NPT7 H9JBD7 A0A1B6EET0 A0A1I8PU56 A0A1E1W3L5 A0A1B6DH86 O97468 A0A0R3P1I8 A0A3B0JKJ8 A0A1B6J4B6 A0A1B6FA14 V5GJJ3 A0A0P5ETW0 K7J2F6 B4MRQ1 A0A1B6MTC6 A0A0P5GIW9 A0A1W4UZX8 A0A212EIJ4 A0A1B6IJ03 A0A1B6DFI4 A0A1W4UMZ9 A0A0A9Z220 A0A0P5H4E6 V5HVY6 A0A0Q9W423 A0A194PK77 A0A0P5G601 A0A2T7PSN6 A0A336M2M6 A0A131ZA78 A0A0P6E442 B3MEM3 A0A023GB93 A0A336MQ24 B4KMD9 B4J632 A0A3B0JD50 B4GCQ9 Q29CQ4 A0A0P5JNR3 T1HYV6 A0A067RFQ3 A0A0Q9W4P3 A0A1B6CB61 A0A224XBX8 A0A069DV00 A0A2J7QU24 A0A0P5GU05 A0A210PJN0 V5GVK5 A0A0K8TS76 A0A0P6DBT1 B4LNX4 A0A3S0ZIL9 A0A195EHV4 F4W8F4 A0A195FB69 A0A158NK58 A0A151X6Y2 A0A0P5GQY4 A0A0B7A069 A0A131Y1K6
A0A194QV33 A0A212FM15 A0A194Q9L2 A0A0L7KN35 A0A2H1VAS8 A0A0L7KS51 A0A0L8HEQ9 A0A0T6BDA3 A0A1Y1N911 A0A2T7PUB6 V9ICK0 A0A1Y1NDZ0 A0A0B7A1I3 A0A0Q9WQA6 D6WYQ8 Q9W1I0 A0A1Y1NCY2 R7TFC3 T1J0V8 A0A194QKQ3 O97469 B4I8U6 B4QB08 A0A1I8PU82 A0A1B6F1X4 B4PA81 E0W1D8 A0A1B6FIW6 A0A1D2MYP4 Q8SXP8 A0A1W4V1I0 B3NPT7 H9JBD7 A0A1B6EET0 A0A1I8PU56 A0A1E1W3L5 A0A1B6DH86 O97468 A0A0R3P1I8 A0A3B0JKJ8 A0A1B6J4B6 A0A1B6FA14 V5GJJ3 A0A0P5ETW0 K7J2F6 B4MRQ1 A0A1B6MTC6 A0A0P5GIW9 A0A1W4UZX8 A0A212EIJ4 A0A1B6IJ03 A0A1B6DFI4 A0A1W4UMZ9 A0A0A9Z220 A0A0P5H4E6 V5HVY6 A0A0Q9W423 A0A194PK77 A0A0P5G601 A0A2T7PSN6 A0A336M2M6 A0A131ZA78 A0A0P6E442 B3MEM3 A0A023GB93 A0A336MQ24 B4KMD9 B4J632 A0A3B0JD50 B4GCQ9 Q29CQ4 A0A0P5JNR3 T1HYV6 A0A067RFQ3 A0A0Q9W4P3 A0A1B6CB61 A0A224XBX8 A0A069DV00 A0A2J7QU24 A0A0P5GU05 A0A210PJN0 V5GVK5 A0A0K8TS76 A0A0P6DBT1 B4LNX4 A0A3S0ZIL9 A0A195EHV4 F4W8F4 A0A195FB69 A0A158NK58 A0A151X6Y2 A0A0P5GQY4 A0A0B7A069 A0A131Y1K6
PDB
4BS2
E-value=2.90642e-61,
Score=595
Ontologies
GO
GO:0003723
GO:0003677
GO:0045292
GO:0000381
GO:0005634
GO:0016607
GO:0008582
GO:1990605
GO:0008345
GO:0048471
GO:0033143
GO:0007417
GO:1900006
GO:0040011
GO:0003729
GO:0007416
GO:0031673
GO:0033119
GO:0045886
GO:0008344
GO:0007274
GO:0007528
GO:0043524
GO:2000331
GO:0007626
GO:0003697
GO:0072553
GO:0007628
GO:0070507
GO:0016021
GO:0000166
GO:0003676
GO:0000154
GO:0008649
GO:0022891
GO:0043565
GO:0016567
GO:0016020
GO:0006260
PANTHER
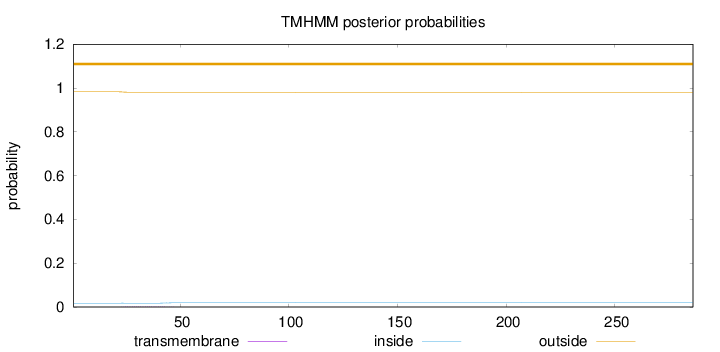
Topology
Length:
286
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04053
Exp number, first 60 AAs:
0.0393
Total prob of N-in:
0.01747
outside
1 - 286
Population Genetic Test Statistics
Pi
13.540588
Theta
14.900509
Tajima's D
-0.181419
CLR
2.184121
CSRT
0.325333733313334
Interpretation
Uncertain
