Pre Gene Modal
BGIBMGA012007
Annotation
PREDICTED:_F-box_only_protein_32_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.711 Nuclear Reliability : 1.273
Sequence
CDS
ATGCCTTTTATTTCAAAAGACTGGCGTTCACCGGGAGAGGAATGGGTGAAAACACAAGAAGGATGGGAAAAGAAGAAAGTGTTGGAGTGCACAGCGCAAAGATACGGTGATATAACTAACTTAAATAACCCGGAAGAAAACCAACAGAAATGGTCCGGTGATGGAGAAGAGGGAGACAAGAGTACAAATGAAGCTGTATCTAGGATACCACCGCACTGTCATATCACGATTAAATGTACTCGAGAAATCGCAGGTTTCAATGGCCTATCCGAAGCCGTCCGTCGTCTAGACTTCTCCTCGGCCGTCCGCGATGTTCGCCGCTTCAATTACATATGCGCTTTGTTGGAGCTATTGCTTCACGGGCAACGACTGACCCACCTGCCCGGGGCTGCTCAGAAACTCTTGCTGTCCATGCTGGAGCAGTTAGCCGACCAGGTGGCGCAGTCGCAACAAAACCTGAACGCTCTCCGCGCTCTGCTGAGCGGGGTGAGTGCGCTGCGCGAGGCCGAGCGGCGGTCATGCTGGGGCCGGCCGCTCGGCTCGAGGGCGCTGTGGGGGCACCACGACCACGCCATAGCCAGGATCCATCACATCGCCAGTGCCATACGTATACAAGAGCCCGGACCCGAGGTTGTGCCGAAACTTCACGATCTACCAGAGGAATGCATTCGGGACATCCTGTTGAGGTTAGCGGATCACAGGGATTTAGATGCAGCGTCGTCAGCATGGAGCGTTATGTCGTCAGTATGCTCGGAGCAGAGGATCTGGCGTGAACTGGTCAGCTTTCACTTCACGAACCAACAGATCGATGCGGCCGTCGCCAAGATCAAGGAGGACGATAAAGAAGTCGAGTGGAAGAAGATCTTCCATCACCTCAGGAAACTGTACGGGCTGCGCGAGGAGGCGCAGTTCGCGGAGACGCTGTCCCTGTGCCGCCACTGCCGCTGCCTTTTTTGGCGCTCGCTCGGACACCCCTGCATTGCCGACCAGTGCCCTGAATACCGCGAGCGCCTCAGAGAAGCTGGAGGACCGTTGCCCCCCTCTCCCGTACCCCCCGCAGCGTTCCTCAAATTCTTCTCGCTCTAA
Protein
MPFISKDWRSPGEEWVKTQEGWEKKKVLECTAQRYGDITNLNNPEENQQKWSGDGEEGDKSTNEAVSRIPPHCHITIKCTREIAGFNGLSEAVRRLDFSSAVRDVRRFNYICALLELLLHGQRLTHLPGAAQKLLLSMLEQLADQVAQSQQNLNALRALLSGVSALREAERRSCWGRPLGSRALWGHHDHAIARIHHIASAIRIQEPGPEVVPKLHDLPEECIRDILLRLADHRDLDAASSAWSVMSSVCSEQRIWRELVSFHFTNQQIDAAVAKIKEDDKEVEWKKIFHHLRKLYGLREEAQFAETLSLCRHCRCLFWRSLGHPCIADQCPEYRERLREAGGPLPPSPVPPAAFLKFFSL
Summary
Uniprot
A0A2A4J9E1
A0A1E1WUC0
A0A194QPM2
A0A212F803
H9JR47
A0A2H1V9X9
+ More
A0A3S2L4M6 A0A0N1PJA4 A0A194Q9F3 A0A1I8JVV5 A0A2M4CSB2 A0A182TK32 A0A2C9GQU3 Q7PCP4 A0A182KT68 A0A182V5F3 A0A1Y1K669 A0A182MR97 A0A2M4ANT3 Q16VJ8 A0A182IZF3 A0A2M4BS88 A0A1Q3G2T0 A0A182Q136 A0A084WM63 D3TNJ6 A0A1A9ULK2 A0A1A9ZLT3 A0A1A9YGT3 A0A1B0BGL4 D6WER2 A0A0L0BKL4 J3JVY1 A0A0K8TPF9 A0A0T6AXT3 T1PDJ6 A0A1I8QCH1 A0A1B0FF40 A0A1Q3G2S7 A0A067R389 A0A2J7QBC5 A0A1B6D9G8 A0A1B6IR81 W5JS13 A0A182S5Q4 E0W101 A0A1B6BX49 A0A2P8YD78 A0A3B0JMR2 B4KUS6 A0A182WD57 B4LCR2 A0A0A1WDE8 A0A1W4VVF4 Q2LZ78 A0A182YC70 A0A0K8V898 A0A034VMC9 B4IZB9 W8BN15 B3M4B1 B3NGZ4 A0A182K2M6 B4HEX5 B4QQA0 Q9VTM4 B4PFN0 A0A1Q3G2E0 A0A182R1Z3 U5EV06 A0A0M4ERG7 A0A182FC20 A0A182P1H9 A0A336M916 X1WIE4 A0A2H8TT21 A0A2S2NFS4 A0A1A9WH58 A0A182U5D1 A0A1W7R8L4 A0A182MXF7 N6UDI7 U4UCC3 B4N4L8 K7J9J5 A0A151WV45 A0A0J7P293 E2ALK3 A0A154PP97 A0A0L7RAL5 T1I227 A0A3L8D3M1 A0A088AP91 A0A2A3EQV6 A0A151HZK4 A0A195EUW0 A0A195CS72 A0A195E369 T1IW22 A0A1W4VVA5
A0A3S2L4M6 A0A0N1PJA4 A0A194Q9F3 A0A1I8JVV5 A0A2M4CSB2 A0A182TK32 A0A2C9GQU3 Q7PCP4 A0A182KT68 A0A182V5F3 A0A1Y1K669 A0A182MR97 A0A2M4ANT3 Q16VJ8 A0A182IZF3 A0A2M4BS88 A0A1Q3G2T0 A0A182Q136 A0A084WM63 D3TNJ6 A0A1A9ULK2 A0A1A9ZLT3 A0A1A9YGT3 A0A1B0BGL4 D6WER2 A0A0L0BKL4 J3JVY1 A0A0K8TPF9 A0A0T6AXT3 T1PDJ6 A0A1I8QCH1 A0A1B0FF40 A0A1Q3G2S7 A0A067R389 A0A2J7QBC5 A0A1B6D9G8 A0A1B6IR81 W5JS13 A0A182S5Q4 E0W101 A0A1B6BX49 A0A2P8YD78 A0A3B0JMR2 B4KUS6 A0A182WD57 B4LCR2 A0A0A1WDE8 A0A1W4VVF4 Q2LZ78 A0A182YC70 A0A0K8V898 A0A034VMC9 B4IZB9 W8BN15 B3M4B1 B3NGZ4 A0A182K2M6 B4HEX5 B4QQA0 Q9VTM4 B4PFN0 A0A1Q3G2E0 A0A182R1Z3 U5EV06 A0A0M4ERG7 A0A182FC20 A0A182P1H9 A0A336M916 X1WIE4 A0A2H8TT21 A0A2S2NFS4 A0A1A9WH58 A0A182U5D1 A0A1W7R8L4 A0A182MXF7 N6UDI7 U4UCC3 B4N4L8 K7J9J5 A0A151WV45 A0A0J7P293 E2ALK3 A0A154PP97 A0A0L7RAL5 T1I227 A0A3L8D3M1 A0A088AP91 A0A2A3EQV6 A0A151HZK4 A0A195EUW0 A0A195CS72 A0A195E369 T1IW22 A0A1W4VVA5
Pubmed
26354079
22118469
19121390
12364791
14747013
17210077
+ More
20966253 28004739 17510324 24438588 20353571 18362917 19820115 26108605 22516182 26369729 25315136 24845553 20920257 23761445 20566863 29403074 17994087 18057021 25830018 15632085 23185243 25244985 25348373 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 23537049 20075255 20798317 30249741
20966253 28004739 17510324 24438588 20353571 18362917 19820115 26108605 22516182 26369729 25315136 24845553 20920257 23761445 20566863 29403074 17994087 18057021 25830018 15632085 23185243 25244985 25348373 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 23537049 20075255 20798317 30249741
EMBL
NWSH01002394
PCG68376.1
GDQN01000476
JAT90578.1
KQ461185
KPJ07483.1
+ More
AGBW02009812 OWR49866.1 BABH01039741 BABH01039742 BABH01039743 BABH01039744 ODYU01001443 SOQ37653.1 RSAL01000157 RVE45593.1 LADJ01048591 KPJ21467.1 KQ459252 KPJ02133.1 GGFL01004054 MBW68232.1 APCN01003267 AAAB01008986 EAA00719.3 GEZM01096060 GEZM01096059 JAV55175.1 AXCM01003002 AXCM01003003 GGFK01008947 MBW42268.1 CH477589 EAT38598.1 GGFJ01006788 MBW55929.1 GFDL01000941 JAV34104.1 AXCN02001699 ATLV01024388 ATLV01024389 KE525351 KFB51307.1 EZ422998 ADD19274.1 JXJN01013888 KQ971326 EFA00885.2 JRES01001711 KNC20625.1 BT127399 AEE62361.1 GDAI01001823 JAI15780.1 LJIG01022553 KRT79964.1 KA646769 KA649811 AFP61398.1 CCAG010005762 GFDL01000920 JAV34125.1 KK852729 KDR17580.1 NEVH01016301 PNF25890.1 GEDC01027095 GEDC01024596 GEDC01014960 JAS10203.1 JAS12702.1 JAS22338.1 GECU01018264 JAS89442.1 ADMH02000596 ETN65715.1 DS235863 EEB19306.1 GEDC01031436 GEDC01021352 JAS05862.1 JAS15946.1 PYGN01000687 PSN42219.1 OUUW01000002 SPP76770.1 CH933809 EDW19332.1 CH940647 EDW70954.1 KRF85210.1 KRF85211.1 GBXI01017273 GBXI01015717 GBXI01009774 JAC97018.1 JAC98574.1 JAD04518.1 CH379069 EAL29630.1 KRT08214.1 KRT08215.1 KRT08216.1 GDHF01017160 GDHF01016996 GDHF01011445 JAI35154.1 JAI35318.1 JAI40869.1 GAKP01015987 GAKP01015986 JAC42965.1 CH916366 EDV96674.1 GAMC01015498 GAMC01015497 GAMC01015496 JAB91059.1 CH902618 EDV39381.2 CH954178 EDV51451.1 KQS43810.1 KQS43811.1 CH480815 EDW41144.1 CM000363 CM002912 EDX10118.1 KMY99051.1 AE014296 AY102690 AAF50023.1 AAM27519.1 CM000159 EDW94179.1 KRK01693.1 KRK01694.1 KRK01695.1 GFDL01001047 JAV33998.1 GANO01002089 JAB57782.1 CP012525 ALC45055.1 UFQT01000715 SSX26715.1 ABLF02025462 ABLF02025469 ABLF02025470 ABLF02025473 ABLF02058292 GFXV01005571 MBW17376.1 GGMR01003360 MBY15979.1 GEHC01000159 JAV47486.1 APGK01027033 KB740648 ENN79765.1 KB632292 ERL91589.1 CH964095 EDW79092.1 AAZX01002696 AAZX01008750 AAZX01012924 KQ982718 KYQ51706.1 LBMM01000327 KMR01213.1 KMR01217.1 GL440609 EFN65705.1 KQ435007 KZC13693.1 KQ414618 KOC67957.1 ACPB03006719 QOIP01000014 RLU14814.1 KZ288193 PBC34070.1 KQ976657 KYM78622.1 KQ981954 KYN32033.1 KQ977329 KYN03495.1 KQ979701 KYN19526.1 JH431604
AGBW02009812 OWR49866.1 BABH01039741 BABH01039742 BABH01039743 BABH01039744 ODYU01001443 SOQ37653.1 RSAL01000157 RVE45593.1 LADJ01048591 KPJ21467.1 KQ459252 KPJ02133.1 GGFL01004054 MBW68232.1 APCN01003267 AAAB01008986 EAA00719.3 GEZM01096060 GEZM01096059 JAV55175.1 AXCM01003002 AXCM01003003 GGFK01008947 MBW42268.1 CH477589 EAT38598.1 GGFJ01006788 MBW55929.1 GFDL01000941 JAV34104.1 AXCN02001699 ATLV01024388 ATLV01024389 KE525351 KFB51307.1 EZ422998 ADD19274.1 JXJN01013888 KQ971326 EFA00885.2 JRES01001711 KNC20625.1 BT127399 AEE62361.1 GDAI01001823 JAI15780.1 LJIG01022553 KRT79964.1 KA646769 KA649811 AFP61398.1 CCAG010005762 GFDL01000920 JAV34125.1 KK852729 KDR17580.1 NEVH01016301 PNF25890.1 GEDC01027095 GEDC01024596 GEDC01014960 JAS10203.1 JAS12702.1 JAS22338.1 GECU01018264 JAS89442.1 ADMH02000596 ETN65715.1 DS235863 EEB19306.1 GEDC01031436 GEDC01021352 JAS05862.1 JAS15946.1 PYGN01000687 PSN42219.1 OUUW01000002 SPP76770.1 CH933809 EDW19332.1 CH940647 EDW70954.1 KRF85210.1 KRF85211.1 GBXI01017273 GBXI01015717 GBXI01009774 JAC97018.1 JAC98574.1 JAD04518.1 CH379069 EAL29630.1 KRT08214.1 KRT08215.1 KRT08216.1 GDHF01017160 GDHF01016996 GDHF01011445 JAI35154.1 JAI35318.1 JAI40869.1 GAKP01015987 GAKP01015986 JAC42965.1 CH916366 EDV96674.1 GAMC01015498 GAMC01015497 GAMC01015496 JAB91059.1 CH902618 EDV39381.2 CH954178 EDV51451.1 KQS43810.1 KQS43811.1 CH480815 EDW41144.1 CM000363 CM002912 EDX10118.1 KMY99051.1 AE014296 AY102690 AAF50023.1 AAM27519.1 CM000159 EDW94179.1 KRK01693.1 KRK01694.1 KRK01695.1 GFDL01001047 JAV33998.1 GANO01002089 JAB57782.1 CP012525 ALC45055.1 UFQT01000715 SSX26715.1 ABLF02025462 ABLF02025469 ABLF02025470 ABLF02025473 ABLF02058292 GFXV01005571 MBW17376.1 GGMR01003360 MBY15979.1 GEHC01000159 JAV47486.1 APGK01027033 KB740648 ENN79765.1 KB632292 ERL91589.1 CH964095 EDW79092.1 AAZX01002696 AAZX01008750 AAZX01012924 KQ982718 KYQ51706.1 LBMM01000327 KMR01213.1 KMR01217.1 GL440609 EFN65705.1 KQ435007 KZC13693.1 KQ414618 KOC67957.1 ACPB03006719 QOIP01000014 RLU14814.1 KZ288193 PBC34070.1 KQ976657 KYM78622.1 KQ981954 KYN32033.1 KQ977329 KYN03495.1 KQ979701 KYN19526.1 JH431604
Proteomes
UP000218220
UP000053240
UP000007151
UP000005204
UP000283053
UP000053268
+ More
UP000076407 UP000075902 UP000075840 UP000007062 UP000075882 UP000075903 UP000075883 UP000008820 UP000075880 UP000075886 UP000030765 UP000078200 UP000092445 UP000092443 UP000092460 UP000007266 UP000037069 UP000095301 UP000095300 UP000092444 UP000027135 UP000235965 UP000000673 UP000075901 UP000009046 UP000245037 UP000268350 UP000009192 UP000075920 UP000008792 UP000192221 UP000001819 UP000076408 UP000001070 UP000007801 UP000008711 UP000075881 UP000001292 UP000000304 UP000000803 UP000002282 UP000075900 UP000092553 UP000069272 UP000075885 UP000007819 UP000091820 UP000075884 UP000019118 UP000030742 UP000007798 UP000002358 UP000075809 UP000036403 UP000000311 UP000076502 UP000053825 UP000015103 UP000279307 UP000005203 UP000242457 UP000078540 UP000078541 UP000078542 UP000078492
UP000076407 UP000075902 UP000075840 UP000007062 UP000075882 UP000075903 UP000075883 UP000008820 UP000075880 UP000075886 UP000030765 UP000078200 UP000092445 UP000092443 UP000092460 UP000007266 UP000037069 UP000095301 UP000095300 UP000092444 UP000027135 UP000235965 UP000000673 UP000075901 UP000009046 UP000245037 UP000268350 UP000009192 UP000075920 UP000008792 UP000192221 UP000001819 UP000076408 UP000001070 UP000007801 UP000008711 UP000075881 UP000001292 UP000000304 UP000000803 UP000002282 UP000075900 UP000092553 UP000069272 UP000075885 UP000007819 UP000091820 UP000075884 UP000019118 UP000030742 UP000007798 UP000002358 UP000075809 UP000036403 UP000000311 UP000076502 UP000053825 UP000015103 UP000279307 UP000005203 UP000242457 UP000078540 UP000078541 UP000078542 UP000078492
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J9E1
A0A1E1WUC0
A0A194QPM2
A0A212F803
H9JR47
A0A2H1V9X9
+ More
A0A3S2L4M6 A0A0N1PJA4 A0A194Q9F3 A0A1I8JVV5 A0A2M4CSB2 A0A182TK32 A0A2C9GQU3 Q7PCP4 A0A182KT68 A0A182V5F3 A0A1Y1K669 A0A182MR97 A0A2M4ANT3 Q16VJ8 A0A182IZF3 A0A2M4BS88 A0A1Q3G2T0 A0A182Q136 A0A084WM63 D3TNJ6 A0A1A9ULK2 A0A1A9ZLT3 A0A1A9YGT3 A0A1B0BGL4 D6WER2 A0A0L0BKL4 J3JVY1 A0A0K8TPF9 A0A0T6AXT3 T1PDJ6 A0A1I8QCH1 A0A1B0FF40 A0A1Q3G2S7 A0A067R389 A0A2J7QBC5 A0A1B6D9G8 A0A1B6IR81 W5JS13 A0A182S5Q4 E0W101 A0A1B6BX49 A0A2P8YD78 A0A3B0JMR2 B4KUS6 A0A182WD57 B4LCR2 A0A0A1WDE8 A0A1W4VVF4 Q2LZ78 A0A182YC70 A0A0K8V898 A0A034VMC9 B4IZB9 W8BN15 B3M4B1 B3NGZ4 A0A182K2M6 B4HEX5 B4QQA0 Q9VTM4 B4PFN0 A0A1Q3G2E0 A0A182R1Z3 U5EV06 A0A0M4ERG7 A0A182FC20 A0A182P1H9 A0A336M916 X1WIE4 A0A2H8TT21 A0A2S2NFS4 A0A1A9WH58 A0A182U5D1 A0A1W7R8L4 A0A182MXF7 N6UDI7 U4UCC3 B4N4L8 K7J9J5 A0A151WV45 A0A0J7P293 E2ALK3 A0A154PP97 A0A0L7RAL5 T1I227 A0A3L8D3M1 A0A088AP91 A0A2A3EQV6 A0A151HZK4 A0A195EUW0 A0A195CS72 A0A195E369 T1IW22 A0A1W4VVA5
A0A3S2L4M6 A0A0N1PJA4 A0A194Q9F3 A0A1I8JVV5 A0A2M4CSB2 A0A182TK32 A0A2C9GQU3 Q7PCP4 A0A182KT68 A0A182V5F3 A0A1Y1K669 A0A182MR97 A0A2M4ANT3 Q16VJ8 A0A182IZF3 A0A2M4BS88 A0A1Q3G2T0 A0A182Q136 A0A084WM63 D3TNJ6 A0A1A9ULK2 A0A1A9ZLT3 A0A1A9YGT3 A0A1B0BGL4 D6WER2 A0A0L0BKL4 J3JVY1 A0A0K8TPF9 A0A0T6AXT3 T1PDJ6 A0A1I8QCH1 A0A1B0FF40 A0A1Q3G2S7 A0A067R389 A0A2J7QBC5 A0A1B6D9G8 A0A1B6IR81 W5JS13 A0A182S5Q4 E0W101 A0A1B6BX49 A0A2P8YD78 A0A3B0JMR2 B4KUS6 A0A182WD57 B4LCR2 A0A0A1WDE8 A0A1W4VVF4 Q2LZ78 A0A182YC70 A0A0K8V898 A0A034VMC9 B4IZB9 W8BN15 B3M4B1 B3NGZ4 A0A182K2M6 B4HEX5 B4QQA0 Q9VTM4 B4PFN0 A0A1Q3G2E0 A0A182R1Z3 U5EV06 A0A0M4ERG7 A0A182FC20 A0A182P1H9 A0A336M916 X1WIE4 A0A2H8TT21 A0A2S2NFS4 A0A1A9WH58 A0A182U5D1 A0A1W7R8L4 A0A182MXF7 N6UDI7 U4UCC3 B4N4L8 K7J9J5 A0A151WV45 A0A0J7P293 E2ALK3 A0A154PP97 A0A0L7RAL5 T1I227 A0A3L8D3M1 A0A088AP91 A0A2A3EQV6 A0A151HZK4 A0A195EUW0 A0A195CS72 A0A195E369 T1IW22 A0A1W4VVA5
Ontologies
GO
PANTHER
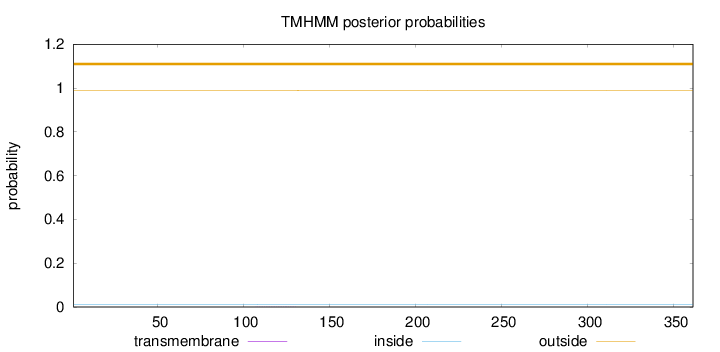
Topology
Length:
361
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01343
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01152
outside
1 - 361
Population Genetic Test Statistics
Pi
21.758059
Theta
20.919014
Tajima's D
0.183917
CLR
1.602432
CSRT
0.428878556072196
Interpretation
Uncertain
