Gene
KWMTBOMO06891
Pre Gene Modal
BGIBMGA012012
Annotation
PREDICTED:_dimethyladenosine_transferase_1?_mitochondrial_[Amyelois_transitella]
Full name
rRNA adenine N(6)-methyltransferase
Location in the cell
Mitochondrial Reliability : 2.119
Sequence
CDS
ATGGCCGTCGCTAAAACTGCATTACAGATAAGGCTGCCTCCACTACCAAGCATCAAGGATGTCATCAAACTCTACAAATTACGCGCTTTACGAGAGCTGTCACAGAATTTTCTCATGGAACCGAGACTTATTGACAAAATAGTGCGTGCCTCAGGTAATATTCAGAATCATACCGTTTGTGAAGTTGGACCCGGCCCGGGTGGAATAACTCGGTCTATAATAAGGCAGGCACCTAAGAAACTTGTTTTAATTGAAAAAGACCCTCGGTTCTTACCATCGTTGGAGCTTTTAGCCGATGCTTGTCGAGATAAAGTCGATGTTGACATTATAACGGGTGATATTTTGAAAACAGATTTAAGTCAATTTATACCGAGTGATGCAAAAGTCCACTGGTTGGATCCACCTCCACCAGTTCACCTCATTGGCAATCTACCATTTAGCGTGTCTACAATTTTGATCATAAGATGGTTGGAGATGATATCTAAACATGAAGGACCATGGTTGTTTGGGAGAACAAGAATGACTCTTACTTTTCAAAAAGAAGTTGCAGAGAGAATGGCTGCCCGTATCTTAGATAAGCAAAGGTGCAGGTTATCAGTTATGTGTCAATCTTGGTGCACTGTAAAATACAATTTTGATATACCAGGTACTGCATTTTTGCCGAAGCCAGATGTAGACGTCGGTGTAGTTACTCTAACACCATTAAAACATCCAATAATAAAATTGCCATTTAAACTTACTGAGAAAGTTGTCCGTCAAATATTTTCAATGAGACAAAAGTATTCGATACGGGGTGCTCAGACGTTGTTCCCAGAAGAAGTGAGAGAAGAGGTAGCTTTAAAAATGTACAACATTGCTGATATTGACCCTGTGACCAGGCCTTTTCAGATAACCAATATGGAATTTGCCAGATTATGTGAAGCATACAATGATATCATAAAGCAACATCCTGAGTTTGAGCATTATGATTACAGAGCACCAAAAAATAAAAATAATCTTATAAATGCAGAGGAAGTTAATATAGAATGTAACATGTGA
Protein
MAVAKTALQIRLPPLPSIKDVIKLYKLRALRELSQNFLMEPRLIDKIVRASGNIQNHTVCEVGPGPGGITRSIIRQAPKKLVLIEKDPRFLPSLELLADACRDKVDVDIITGDILKTDLSQFIPSDAKVHWLDPPPPVHLIGNLPFSVSTILIIRWLEMISKHEGPWLFGRTRMTLTFQKEVAERMAARILDKQRCRLSVMCQSWCTVKYNFDIPGTAFLPKPDVDVGVVTLTPLKHPIIKLPFKLTEKVVRQIFSMRQKYSIRGAQTLFPEEVREEVALKMYNIADIDPVTRPFQITNMEFARLCEAYNDIIKQHPEFEHYDYRAPKNKNNLINAEEVNIECNM
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family.
Uniprot
H9JR52
A0A2H1W125
A0A0L7KTT3
A0A194QV99
I4DKF4
A0A2A4J8C7
+ More
A0A194QF91 A0A212F7Z9 S4PK95 A0A1Y1KK19 D6W6C5 A0A2S2N8S7 A0A067RF29 A0A2H8TQG3 A0A2P8YBT2 A0A336L5W5 A0A1W4XU91 A0A2S2QGN5 B0WLE5 A0A1Q3FIT8 Q16VK0 A0A2J7QBC4 A0A1B0CTH1 A0A182GSM7 A0A182H732 T1HMC6 A0A182X039 A0A182WD60 A0A182KCD3 A0A182M861 A0A182R1Z1 A0A182KT50 A0A182VLF7 A0A182P1I2 A0A084WM64 A0A182HPN9 Q7PZX0 U4U6N5 A0A2M4A9Q5 A0A2M4AA80 A0A2M4AA67 A0A182UIE6 A0A182IU38 A0A182MXF5 A0A1B3PDG1 A0A182Q3Q8 A0A171AGT9 A0A2M4BSP9 A0A026WBF3 W5JMW0 A0A1B3PDA8 A0A2M4BSR4 A0A2M4A9Z2 A0A0J7NU55 A0A0A9WG27 A0A2M4BSR5 A0A2M3Z5Z7 A0A2M3Z5X7 A0A2M3ZHE5 A0A0K8SDC9 E2A745 A0A154PPD3 A0A1J1IMQ0 A0A1B0CWR3 E9FZG5 E9IE08 A0A0Q3PGA9 A0A1B6DY39 K4FU19 A0A091JTD3 F4WZE1 A0A151INV7 A0A195E5N8 A0A091UI01 A0A091MZD5 A0A2I0MVE0 A0A093GRD6 A0A091JL61 A0A158NG55 A0A091I704 A0A1V4JMF9 A0A088AQJ2 A0A094LDR0 G1KDU6 A0A0P4W523 H3AXD8 A0A091WAW2 A0A0L7RE23 A0A2A3EKP1 A0A151X248 A0A091VV95 A0A3B4BLJ4 A0A195ETD9 A0A093QNR6 A0A0C9Q9U8 A0A093ID92 K7FGX5 F6W4I0 A0A091FJS5 A0A091KXT4
A0A194QF91 A0A212F7Z9 S4PK95 A0A1Y1KK19 D6W6C5 A0A2S2N8S7 A0A067RF29 A0A2H8TQG3 A0A2P8YBT2 A0A336L5W5 A0A1W4XU91 A0A2S2QGN5 B0WLE5 A0A1Q3FIT8 Q16VK0 A0A2J7QBC4 A0A1B0CTH1 A0A182GSM7 A0A182H732 T1HMC6 A0A182X039 A0A182WD60 A0A182KCD3 A0A182M861 A0A182R1Z1 A0A182KT50 A0A182VLF7 A0A182P1I2 A0A084WM64 A0A182HPN9 Q7PZX0 U4U6N5 A0A2M4A9Q5 A0A2M4AA80 A0A2M4AA67 A0A182UIE6 A0A182IU38 A0A182MXF5 A0A1B3PDG1 A0A182Q3Q8 A0A171AGT9 A0A2M4BSP9 A0A026WBF3 W5JMW0 A0A1B3PDA8 A0A2M4BSR4 A0A2M4A9Z2 A0A0J7NU55 A0A0A9WG27 A0A2M4BSR5 A0A2M3Z5Z7 A0A2M3Z5X7 A0A2M3ZHE5 A0A0K8SDC9 E2A745 A0A154PPD3 A0A1J1IMQ0 A0A1B0CWR3 E9FZG5 E9IE08 A0A0Q3PGA9 A0A1B6DY39 K4FU19 A0A091JTD3 F4WZE1 A0A151INV7 A0A195E5N8 A0A091UI01 A0A091MZD5 A0A2I0MVE0 A0A093GRD6 A0A091JL61 A0A158NG55 A0A091I704 A0A1V4JMF9 A0A088AQJ2 A0A094LDR0 G1KDU6 A0A0P4W523 H3AXD8 A0A091WAW2 A0A0L7RE23 A0A2A3EKP1 A0A151X248 A0A091VV95 A0A3B4BLJ4 A0A195ETD9 A0A093QNR6 A0A0C9Q9U8 A0A093ID92 K7FGX5 F6W4I0 A0A091FJS5 A0A091KXT4
EC Number
2.1.1.-
Pubmed
19121390
26227816
26354079
22651552
22118469
23622113
+ More
28004739 18362917 19820115 24845553 29403074 17510324 26483478 20966253 24438588 12364791 23537049 24508170 30249741 20920257 23761445 25401762 26823975 20798317 21292972 21282665 23056606 21719571 23371554 21347285 9215903 17381049 19892987
28004739 18362917 19820115 24845553 29403074 17510324 26483478 20966253 24438588 12364791 23537049 24508170 30249741 20920257 23761445 25401762 26823975 20798317 21292972 21282665 23056606 21719571 23371554 21347285 9215903 17381049 19892987
EMBL
BABH01039748
ODYU01005400
SOQ46194.1
JTDY01005756
KOB66668.1
KQ461185
+ More
KPJ07481.1 AK401772 BAM18394.1 NWSH01002394 PCG68377.1 KQ459252 KPJ02136.1 AGBW02009812 OWR49867.1 GAIX01004625 JAA87935.1 GEZM01086634 JAV59137.1 KQ971319 EFA11369.2 GGMR01000879 MBY13498.1 KK852729 KDR17578.1 GFXV01004425 GFXV01006087 GFXV01007923 MBW16230.1 MBW17892.1 MBW19728.1 PYGN01000721 PSN41722.1 UFQS01001716 UFQT01001716 SSX11954.1 SSX31501.1 GGMS01007658 MBY76861.1 DS231984 EDS30438.1 GFDL01007611 JAV27434.1 CH477589 EAT38596.1 NEVH01016301 PNF25888.1 AJWK01027598 JXUM01017272 KQ560479 KXJ82209.1 JXUM01027871 JXUM01027872 JXUM01027873 JXUM01027874 ACPB03004195 AXCM01003002 ATLV01024389 KE525351 KFB51308.1 APCN01003267 AAAB01008986 EAA00207.3 KB632095 ERL88732.1 GGFK01004139 MBW37460.1 GGFK01004358 MBW37679.1 GGFK01004366 MBW37687.1 KU659967 AOG17765.1 AXCN02001699 GEMB01001083 JAS02064.1 GGFJ01006891 MBW56032.1 KK107346 QOIP01000007 EZA52374.1 RLU20234.1 ADMH02000596 ETN65717.1 KU659914 AOG17713.1 GGFJ01006893 MBW56034.1 GGFK01004303 MBW37624.1 LBMM01001652 KMQ95940.1 GBHO01039789 GDHC01019280 GDHC01017602 JAG03815.1 JAP99348.1 JAQ01027.1 GGFJ01006892 MBW56033.1 GGFM01003192 MBW23943.1 GGFM01003186 MBW23937.1 GGFM01007144 MBW27895.1 GBRD01014528 JAG51298.1 GL437252 EFN70777.1 KQ434981 KZC13118.1 CVRI01000055 CRL01018.1 AJWK01032773 GL732528 EFX87273.1 GL762556 EFZ21203.1 LMAW01002567 KQK79779.1 GEDC01006710 JAS30588.1 JX053097 AFK11325.1 KK532385 KFP28417.1 GL888479 EGI60385.1 KQ976932 KYN07028.1 KQ979609 KYN20207.1 KK434327 KFQ89807.1 KL373376 KFP82047.1 AKCR02000002 PKK33641.1 KL217019 KFV72848.1 KK502094 KFP20475.1 ADTU01001550 KL218240 KFP03253.1 LSYS01006902 OPJ72937.1 KL289797 KFZ69438.1 GDRN01101012 JAI58455.1 AFYH01150630 AFYH01150631 AFYH01150632 AFYH01150633 AFYH01150634 KK735008 KFR11898.1 KQ414611 KOC69227.1 KZ288227 PBC31822.1 KQ982585 KYQ54336.1 KL410062 KFQ93593.1 KQ981979 KYN31426.1 KL425443 KFW90161.1 GBYB01011153 JAG80920.1 KK585404 KFV96733.1 AGCU01032451 AGCU01032452 AGCU01032453 AGCU01032454 AGCU01032455 KL447196 KFO70755.1 KK758363 KFP44080.1
KPJ07481.1 AK401772 BAM18394.1 NWSH01002394 PCG68377.1 KQ459252 KPJ02136.1 AGBW02009812 OWR49867.1 GAIX01004625 JAA87935.1 GEZM01086634 JAV59137.1 KQ971319 EFA11369.2 GGMR01000879 MBY13498.1 KK852729 KDR17578.1 GFXV01004425 GFXV01006087 GFXV01007923 MBW16230.1 MBW17892.1 MBW19728.1 PYGN01000721 PSN41722.1 UFQS01001716 UFQT01001716 SSX11954.1 SSX31501.1 GGMS01007658 MBY76861.1 DS231984 EDS30438.1 GFDL01007611 JAV27434.1 CH477589 EAT38596.1 NEVH01016301 PNF25888.1 AJWK01027598 JXUM01017272 KQ560479 KXJ82209.1 JXUM01027871 JXUM01027872 JXUM01027873 JXUM01027874 ACPB03004195 AXCM01003002 ATLV01024389 KE525351 KFB51308.1 APCN01003267 AAAB01008986 EAA00207.3 KB632095 ERL88732.1 GGFK01004139 MBW37460.1 GGFK01004358 MBW37679.1 GGFK01004366 MBW37687.1 KU659967 AOG17765.1 AXCN02001699 GEMB01001083 JAS02064.1 GGFJ01006891 MBW56032.1 KK107346 QOIP01000007 EZA52374.1 RLU20234.1 ADMH02000596 ETN65717.1 KU659914 AOG17713.1 GGFJ01006893 MBW56034.1 GGFK01004303 MBW37624.1 LBMM01001652 KMQ95940.1 GBHO01039789 GDHC01019280 GDHC01017602 JAG03815.1 JAP99348.1 JAQ01027.1 GGFJ01006892 MBW56033.1 GGFM01003192 MBW23943.1 GGFM01003186 MBW23937.1 GGFM01007144 MBW27895.1 GBRD01014528 JAG51298.1 GL437252 EFN70777.1 KQ434981 KZC13118.1 CVRI01000055 CRL01018.1 AJWK01032773 GL732528 EFX87273.1 GL762556 EFZ21203.1 LMAW01002567 KQK79779.1 GEDC01006710 JAS30588.1 JX053097 AFK11325.1 KK532385 KFP28417.1 GL888479 EGI60385.1 KQ976932 KYN07028.1 KQ979609 KYN20207.1 KK434327 KFQ89807.1 KL373376 KFP82047.1 AKCR02000002 PKK33641.1 KL217019 KFV72848.1 KK502094 KFP20475.1 ADTU01001550 KL218240 KFP03253.1 LSYS01006902 OPJ72937.1 KL289797 KFZ69438.1 GDRN01101012 JAI58455.1 AFYH01150630 AFYH01150631 AFYH01150632 AFYH01150633 AFYH01150634 KK735008 KFR11898.1 KQ414611 KOC69227.1 KZ288227 PBC31822.1 KQ982585 KYQ54336.1 KL410062 KFQ93593.1 KQ981979 KYN31426.1 KL425443 KFW90161.1 GBYB01011153 JAG80920.1 KK585404 KFV96733.1 AGCU01032451 AGCU01032452 AGCU01032453 AGCU01032454 AGCU01032455 KL447196 KFO70755.1 KK758363 KFP44080.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000218220
UP000053268
UP000007151
+ More
UP000007266 UP000027135 UP000245037 UP000192223 UP000002320 UP000008820 UP000235965 UP000092461 UP000069940 UP000249989 UP000015103 UP000076407 UP000075920 UP000075881 UP000075883 UP000075900 UP000075882 UP000075903 UP000075885 UP000030765 UP000075840 UP000007062 UP000030742 UP000075902 UP000075880 UP000075884 UP000075886 UP000053097 UP000279307 UP000000673 UP000036403 UP000000311 UP000076502 UP000183832 UP000000305 UP000051836 UP000007755 UP000078542 UP000078492 UP000053872 UP000053875 UP000053119 UP000005205 UP000054308 UP000190648 UP000005203 UP000001646 UP000008672 UP000053605 UP000053825 UP000242457 UP000075809 UP000053283 UP000261440 UP000078541 UP000007267 UP000002281 UP000053760
UP000007266 UP000027135 UP000245037 UP000192223 UP000002320 UP000008820 UP000235965 UP000092461 UP000069940 UP000249989 UP000015103 UP000076407 UP000075920 UP000075881 UP000075883 UP000075900 UP000075882 UP000075903 UP000075885 UP000030765 UP000075840 UP000007062 UP000030742 UP000075902 UP000075880 UP000075884 UP000075886 UP000053097 UP000279307 UP000000673 UP000036403 UP000000311 UP000076502 UP000183832 UP000000305 UP000051836 UP000007755 UP000078542 UP000078492 UP000053872 UP000053875 UP000053119 UP000005205 UP000054308 UP000190648 UP000005203 UP000001646 UP000008672 UP000053605 UP000053825 UP000242457 UP000075809 UP000053283 UP000261440 UP000078541 UP000007267 UP000002281 UP000053760
PRIDE
Interpro
IPR029063
SAM-dependent_MTases
+ More
IPR001737 KsgA/Erm
IPR023165 rRNA_Ade_diMease-like
IPR011530 rRNA_adenine_dimethylase
IPR020598 rRNA_Ade_methylase_Trfase_N
IPR020596 rRNA_Ade_Mease_Trfase_CS
IPR018628 Coa3_cc
IPR011876 IsopentenylPP_isomerase_typ1
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR000086 NUDIX_hydrolase_dom
IPR001737 KsgA/Erm
IPR023165 rRNA_Ade_diMease-like
IPR011530 rRNA_adenine_dimethylase
IPR020598 rRNA_Ade_methylase_Trfase_N
IPR020596 rRNA_Ade_Mease_Trfase_CS
IPR018628 Coa3_cc
IPR011876 IsopentenylPP_isomerase_typ1
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR000086 NUDIX_hydrolase_dom
Gene 3D
ProteinModelPortal
H9JR52
A0A2H1W125
A0A0L7KTT3
A0A194QV99
I4DKF4
A0A2A4J8C7
+ More
A0A194QF91 A0A212F7Z9 S4PK95 A0A1Y1KK19 D6W6C5 A0A2S2N8S7 A0A067RF29 A0A2H8TQG3 A0A2P8YBT2 A0A336L5W5 A0A1W4XU91 A0A2S2QGN5 B0WLE5 A0A1Q3FIT8 Q16VK0 A0A2J7QBC4 A0A1B0CTH1 A0A182GSM7 A0A182H732 T1HMC6 A0A182X039 A0A182WD60 A0A182KCD3 A0A182M861 A0A182R1Z1 A0A182KT50 A0A182VLF7 A0A182P1I2 A0A084WM64 A0A182HPN9 Q7PZX0 U4U6N5 A0A2M4A9Q5 A0A2M4AA80 A0A2M4AA67 A0A182UIE6 A0A182IU38 A0A182MXF5 A0A1B3PDG1 A0A182Q3Q8 A0A171AGT9 A0A2M4BSP9 A0A026WBF3 W5JMW0 A0A1B3PDA8 A0A2M4BSR4 A0A2M4A9Z2 A0A0J7NU55 A0A0A9WG27 A0A2M4BSR5 A0A2M3Z5Z7 A0A2M3Z5X7 A0A2M3ZHE5 A0A0K8SDC9 E2A745 A0A154PPD3 A0A1J1IMQ0 A0A1B0CWR3 E9FZG5 E9IE08 A0A0Q3PGA9 A0A1B6DY39 K4FU19 A0A091JTD3 F4WZE1 A0A151INV7 A0A195E5N8 A0A091UI01 A0A091MZD5 A0A2I0MVE0 A0A093GRD6 A0A091JL61 A0A158NG55 A0A091I704 A0A1V4JMF9 A0A088AQJ2 A0A094LDR0 G1KDU6 A0A0P4W523 H3AXD8 A0A091WAW2 A0A0L7RE23 A0A2A3EKP1 A0A151X248 A0A091VV95 A0A3B4BLJ4 A0A195ETD9 A0A093QNR6 A0A0C9Q9U8 A0A093ID92 K7FGX5 F6W4I0 A0A091FJS5 A0A091KXT4
A0A194QF91 A0A212F7Z9 S4PK95 A0A1Y1KK19 D6W6C5 A0A2S2N8S7 A0A067RF29 A0A2H8TQG3 A0A2P8YBT2 A0A336L5W5 A0A1W4XU91 A0A2S2QGN5 B0WLE5 A0A1Q3FIT8 Q16VK0 A0A2J7QBC4 A0A1B0CTH1 A0A182GSM7 A0A182H732 T1HMC6 A0A182X039 A0A182WD60 A0A182KCD3 A0A182M861 A0A182R1Z1 A0A182KT50 A0A182VLF7 A0A182P1I2 A0A084WM64 A0A182HPN9 Q7PZX0 U4U6N5 A0A2M4A9Q5 A0A2M4AA80 A0A2M4AA67 A0A182UIE6 A0A182IU38 A0A182MXF5 A0A1B3PDG1 A0A182Q3Q8 A0A171AGT9 A0A2M4BSP9 A0A026WBF3 W5JMW0 A0A1B3PDA8 A0A2M4BSR4 A0A2M4A9Z2 A0A0J7NU55 A0A0A9WG27 A0A2M4BSR5 A0A2M3Z5Z7 A0A2M3Z5X7 A0A2M3ZHE5 A0A0K8SDC9 E2A745 A0A154PPD3 A0A1J1IMQ0 A0A1B0CWR3 E9FZG5 E9IE08 A0A0Q3PGA9 A0A1B6DY39 K4FU19 A0A091JTD3 F4WZE1 A0A151INV7 A0A195E5N8 A0A091UI01 A0A091MZD5 A0A2I0MVE0 A0A093GRD6 A0A091JL61 A0A158NG55 A0A091I704 A0A1V4JMF9 A0A088AQJ2 A0A094LDR0 G1KDU6 A0A0P4W523 H3AXD8 A0A091WAW2 A0A0L7RE23 A0A2A3EKP1 A0A151X248 A0A091VV95 A0A3B4BLJ4 A0A195ETD9 A0A093QNR6 A0A0C9Q9U8 A0A093ID92 K7FGX5 F6W4I0 A0A091FJS5 A0A091KXT4
PDB
4GC9
E-value=1.50868e-91,
Score=857
Ontologies
GO
PANTHER
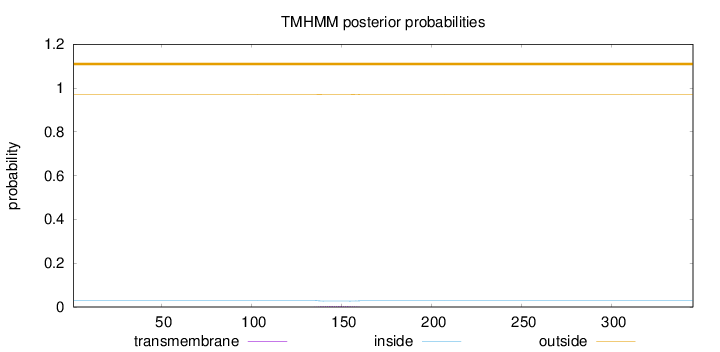
Topology
Length:
345
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0752199999999998
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.02931
outside
1 - 345
Population Genetic Test Statistics
Pi
17.838208
Theta
13.787819
Tajima's D
0.900547
CLR
1.359327
CSRT
0.635418229088546
Interpretation
Uncertain
