Gene
KWMTBOMO06890
Pre Gene Modal
BGIBMGA012005
Annotation
PREDICTED:_tRNA_dimethylallyltransferase?_mitochondrial-like_[Bombyx_mori]
Full name
tRNA dimethylallyltransferase
Alternative Name
Isopentenyl-diphosphate:tRNA isopentenyltransferase
hGRO1
tRNA isopentenyltransferase 1
tRNA isopentenyltransferase
hGRO1
tRNA isopentenyltransferase 1
tRNA isopentenyltransferase
Location in the cell
Nuclear Reliability : 4.452
Sequence
CDS
ATGGCTACTTTTATCTTGTTTCAGGATGTCCACGATAAGCGTCTTGACGCTCGCGTCGACTCGATGCTGGAAGCGGGCCTCATTGAGGAGCTGCTGGATTTCCACGAGAGGCACAACAGGCACCGGATACAGGACGGGAGCTCACCAGACTACACCAAAGGCGTTTTCCAAACTCTTGGCTTCAAAGAATTCCACGATTACCTGATGATGTCCGAAGACGAAAGGAATTCGGAGGCGGGCAAGAAACTACTGAAGACCAGCATTGAAAACATGAAGATGGCCACCCGGAGATACGCGAGACGTCAAAACAAAATGTTTCGTCAAAGATTTTTAGAACATCCCAGAAGAGAGGTGCCTTTAGTTTACGAATTGGACACTACAGATGTAACCAAATGGGATGAAAACGTGAAAAACAAGGCTATTAAAATAATAGAAAGTTTCATCAATAGCACTGATTCTGATGTTTTACCAATAAAACAGCATCTTCAAAGTGATAAACTGCATACAGACGGAAATTCTTTTAATTTTTGTGAAATTTGTAATCGAATCATAATAGGCGATAACGTTTATGCTATTCATTTAAAATCTGTGCGTCACATGAAGGTTTTAAAGAAAATGAGATCACAAAACAAGGAACAAAAACAAGATTAA
Protein
MATFILFQDVHDKRLDARVDSMLEAGLIEELLDFHERHNRHRIQDGSSPDYTKGVFQTLGFKEFHDYLMMSEDERNSEAGKKLLKTSIENMKMATRRYARRQNKMFRQRFLEHPRREVPLVYELDTTDVTKWDENVKNKAIKIIESFINSTDSDVLPIKQHLQSDKLHTDGNSFNFCEICNRIIIGDNVYAIHLKSVRHMKVLKKMRSQNKEQKQD
Summary
Description
Catalyzes the transfer of a dimethylallyl group onto the adenine at position 37.
Catalyzes the transfer of a dimethylallyl group onto the adenine at position 37 of both cytosolic and mitochondrial tRNAs, leading to the formation of N6-(dimethylallyl)adenosine (i(6)A).
Catalyzes the transfer of a dimethylallyl group onto the adenine at position 37 of both cytosolic and mitochondrial tRNAs, leading to the formation of N6-(dimethylallyl)adenosine (i(6)A).
Catalytic Activity
adenosine(37) in tRNA + dimethylallyl diphosphate = diphosphate + N(6)-dimethylallyladenosine(37) in tRNA
Similarity
Belongs to the IPP transferase family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Disease mutation
Metal-binding
Mitochondrion
Nucleotide-binding
Phosphoprotein
Polymorphism
Primary mitochondrial disease
Reference proteome
Transferase
Transit peptide
tRNA processing
Zinc
Zinc-finger
Nucleus
Feature
chain tRNA dimethylallyltransferase
splice variant In isoform 2 and isoform 3.
sequence variant In COXPD35.
splice variant In isoform 2 and isoform 3.
sequence variant In COXPD35.
Uniprot
A0A2H1VZD8
A0A2A4J9M1
A0A194QR76
A0A194Q9E8
K7J5P2
D6WDB1
+ More
A0A154PPN4 A0A3L8DGV1 A0A212F819 A0A151IXH3 A0A195CY49 A0A087ZX87 A0A1B6E6L8 A0A224XJA2 A0A1W4XCH3 A0A069DQG6 A0A067R5L3 A0A151XH91 A0A158P2W8 N6TRD4 A0A3R7MJU5 A0A023FXU1 A0A1Y1M5B9 A0A0C9PRS7 U4U1W4 A0A2A3EI31 A0A2R5L5Q2 A0A195EZX8 A0A2I3LRI4 A0A3Q7X4X4 A0A2K6B163 A0A2K5U7C2 F7FQY3 A0A2J7QBD1 A0A2K5Z319 A0A384DT44 F6XBX7 A0A0D9S7N8 A0A2K5DYS2 A0A096MMW7 A0A2K5KC61 A0A2K5DYZ9 Q9H3H1-2 A0A2J8Y697 A0A3Q7XYH3 Q80UN9-2 A0A2K6B154 F7GJT8 I7G8M2 A0A2I2Z503 A0A2F0AWM5 H9F4Y3 G7NUC5 G7MG98 Q9H3H1-3 M3W562 A0A2K5RFZ3 A0A2K5Z325 A0A2K5DZ25 Q3T7C5 A0A2R8ZLW7 A0A2I3RL05 A0A2Y9J4F2 A0A1S3WGH9 G1LCG9 A0A3Q7P9Y3 G3HN83 D2HXL8 A0A2I3HS54 H9G668 A0A384DTX5 Q9H3H1-5 U3FSI0 M7BJR6 G1SGQ1 A0A1E1X3E8 A0A2K5KC58 H2N7V3 A0A1Z5L5F8 A0A2Y9IZV5 G3R3K0 A0A2K5DYN8 A0A2J8Y6C7 A0A3Q0DFI2 Q3T7C7 A0A293LL16 A0A2K5RFT6 A0A2K6EWL2 A0A1S3F8M3 A0A2R8ZPL5 H2PYR3 G1RN30 Q53F11 A0A1B6EHY5 Q9H3H1 A0A1Y1M5G9 M3YS73 A0A2K6MUL4
A0A154PPN4 A0A3L8DGV1 A0A212F819 A0A151IXH3 A0A195CY49 A0A087ZX87 A0A1B6E6L8 A0A224XJA2 A0A1W4XCH3 A0A069DQG6 A0A067R5L3 A0A151XH91 A0A158P2W8 N6TRD4 A0A3R7MJU5 A0A023FXU1 A0A1Y1M5B9 A0A0C9PRS7 U4U1W4 A0A2A3EI31 A0A2R5L5Q2 A0A195EZX8 A0A2I3LRI4 A0A3Q7X4X4 A0A2K6B163 A0A2K5U7C2 F7FQY3 A0A2J7QBD1 A0A2K5Z319 A0A384DT44 F6XBX7 A0A0D9S7N8 A0A2K5DYS2 A0A096MMW7 A0A2K5KC61 A0A2K5DYZ9 Q9H3H1-2 A0A2J8Y697 A0A3Q7XYH3 Q80UN9-2 A0A2K6B154 F7GJT8 I7G8M2 A0A2I2Z503 A0A2F0AWM5 H9F4Y3 G7NUC5 G7MG98 Q9H3H1-3 M3W562 A0A2K5RFZ3 A0A2K5Z325 A0A2K5DZ25 Q3T7C5 A0A2R8ZLW7 A0A2I3RL05 A0A2Y9J4F2 A0A1S3WGH9 G1LCG9 A0A3Q7P9Y3 G3HN83 D2HXL8 A0A2I3HS54 H9G668 A0A384DTX5 Q9H3H1-5 U3FSI0 M7BJR6 G1SGQ1 A0A1E1X3E8 A0A2K5KC58 H2N7V3 A0A1Z5L5F8 A0A2Y9IZV5 G3R3K0 A0A2K5DYN8 A0A2J8Y6C7 A0A3Q0DFI2 Q3T7C7 A0A293LL16 A0A2K5RFT6 A0A2K6EWL2 A0A1S3F8M3 A0A2R8ZPL5 H2PYR3 G1RN30 Q53F11 A0A1B6EHY5 Q9H3H1 A0A1Y1M5G9 M3YS73 A0A2K6MUL4
EC Number
2.5.1.75
Pubmed
26354079
20075255
18362917
19820115
30249741
22118469
+ More
26334808 24845553 21347285 23537049 28004739 17431167 11111046 15870694 14702039 16710414 15489334 11560893 17081983 20068231 21269460 21406692 23186163 24901367 28185376 16141072 19468303 17194215 22398555 25319552 22002653 17975172 22722832 16136131 20010809 21804562 21881562 25243066 23624526 21993624 28503490 28528879 8125298 9373149
26334808 24845553 21347285 23537049 28004739 17431167 11111046 15870694 14702039 16710414 15489334 11560893 17081983 20068231 21269460 21406692 23186163 24901367 28185376 16141072 19468303 17194215 22398555 25319552 22002653 17975172 22722832 16136131 20010809 21804562 21881562 25243066 23624526 21993624 28503490 28528879 8125298 9373149
EMBL
ODYU01005400
SOQ46195.1
NWSH01002394
PCG68378.1
KQ461185
KPJ07480.1
+ More
KQ459252 KPJ02137.1 KQ971321 EEZ99485.2 KQ435012 KZC13841.1 QOIP01000009 RLU19149.1 AGBW02009812 OWR49868.1 KQ980801 KYN12801.1 KQ977185 KYN05074.1 GEDC01003753 JAS33545.1 GFTR01003860 JAW12566.1 GBGD01002739 JAC86150.1 KK852729 KDR17577.1 KQ982130 KYQ59727.1 ADTU01007582 ADTU01007583 ADTU01007584 ADTU01007585 ADTU01007586 ADTU01007587 ADTU01007588 ADTU01007589 ADTU01007590 ADTU01007591 APGK01057365 KB741280 ENN70881.1 QCYY01000659 ROT83645.1 GBBL01001782 JAC25538.1 GEZM01040044 GEZM01040042 JAV80972.1 GBYB01004068 JAG73835.1 KB631604 ERL84601.1 KZ288237 PBC31377.1 GGLE01000692 MBY04818.1 KQ981905 KYN33429.1 AHZZ02015817 AHZZ02015818 AQIA01004583 AQIA01004584 AQIA01004585 AQIA01004586 AQIA01004587 JSUE03001184 JSUE03001185 NEVH01016301 PNF25887.1 AQIB01144671 AQIB01144672 AQIB01144673 AQIB01144674 AF074918 AY702944 AY702945 AY303390 AK000068 CR457226 AL033527 BC010741 BC107569 BC128155 AY052768 NDHI03003278 PNJ89802.1 AK003556 AL606906 BC019812 BC051040 AB170647 BAE87710.1 CABD030002601 CABD030002602 CABD030002603 NTJE010014569 MBV95139.1 JU325936 AFE69692.1 CM001276 EHH49804.1 CM001253 EHH14644.1 AANG04003279 AY702935 AAW63396.1 AJFE02051312 AJFE02051313 AJFE02051314 AJFE02051315 AJFE02051316 AJFE02051317 AJFE02051318 AJFE02051319 AACZ04033017 NBAG03000275 PNI52424.1 ACTA01098509 JH000537 EGV92584.1 GL193633 EFB13518.1 ADFV01076468 ADFV01076469 AAWZ02028779 AAWZ02028780 GAMS01009588 GAMR01008134 GAMP01002514 JAB13548.1 JAB25798.1 JAB50241.1 KB563787 EMP28447.1 AAGW02003112 AAGW02003113 GFAC01005403 JAT93785.1 ABGA01074454 ABGA01074455 PNJ89796.1 GFJQ02004317 JAW02653.1 PNJ89801.1 AY702933 AAW63394.1 GFWV01003791 MAA28521.1 GABE01011185 JAA33554.1 PNI52422.1 AK223478 BAD97198.1 GECZ01032222 JAS37547.1 GEZM01040043 JAV80973.1 AEYP01007632 AEYP01007633 AEYP01007634 AEYP01007635
KQ459252 KPJ02137.1 KQ971321 EEZ99485.2 KQ435012 KZC13841.1 QOIP01000009 RLU19149.1 AGBW02009812 OWR49868.1 KQ980801 KYN12801.1 KQ977185 KYN05074.1 GEDC01003753 JAS33545.1 GFTR01003860 JAW12566.1 GBGD01002739 JAC86150.1 KK852729 KDR17577.1 KQ982130 KYQ59727.1 ADTU01007582 ADTU01007583 ADTU01007584 ADTU01007585 ADTU01007586 ADTU01007587 ADTU01007588 ADTU01007589 ADTU01007590 ADTU01007591 APGK01057365 KB741280 ENN70881.1 QCYY01000659 ROT83645.1 GBBL01001782 JAC25538.1 GEZM01040044 GEZM01040042 JAV80972.1 GBYB01004068 JAG73835.1 KB631604 ERL84601.1 KZ288237 PBC31377.1 GGLE01000692 MBY04818.1 KQ981905 KYN33429.1 AHZZ02015817 AHZZ02015818 AQIA01004583 AQIA01004584 AQIA01004585 AQIA01004586 AQIA01004587 JSUE03001184 JSUE03001185 NEVH01016301 PNF25887.1 AQIB01144671 AQIB01144672 AQIB01144673 AQIB01144674 AF074918 AY702944 AY702945 AY303390 AK000068 CR457226 AL033527 BC010741 BC107569 BC128155 AY052768 NDHI03003278 PNJ89802.1 AK003556 AL606906 BC019812 BC051040 AB170647 BAE87710.1 CABD030002601 CABD030002602 CABD030002603 NTJE010014569 MBV95139.1 JU325936 AFE69692.1 CM001276 EHH49804.1 CM001253 EHH14644.1 AANG04003279 AY702935 AAW63396.1 AJFE02051312 AJFE02051313 AJFE02051314 AJFE02051315 AJFE02051316 AJFE02051317 AJFE02051318 AJFE02051319 AACZ04033017 NBAG03000275 PNI52424.1 ACTA01098509 JH000537 EGV92584.1 GL193633 EFB13518.1 ADFV01076468 ADFV01076469 AAWZ02028779 AAWZ02028780 GAMS01009588 GAMR01008134 GAMP01002514 JAB13548.1 JAB25798.1 JAB50241.1 KB563787 EMP28447.1 AAGW02003112 AAGW02003113 GFAC01005403 JAT93785.1 ABGA01074454 ABGA01074455 PNJ89796.1 GFJQ02004317 JAW02653.1 PNJ89801.1 AY702933 AAW63394.1 GFWV01003791 MAA28521.1 GABE01011185 JAA33554.1 PNI52422.1 AK223478 BAD97198.1 GECZ01032222 JAS37547.1 GEZM01040043 JAV80973.1 AEYP01007632 AEYP01007633 AEYP01007634 AEYP01007635
Proteomes
UP000218220
UP000053240
UP000053268
UP000002358
UP000007266
UP000076502
+ More
UP000279307 UP000007151 UP000078492 UP000078542 UP000005203 UP000192223 UP000027135 UP000075809 UP000005205 UP000019118 UP000283509 UP000030742 UP000242457 UP000078541 UP000028761 UP000286642 UP000233120 UP000233100 UP000006718 UP000235965 UP000233140 UP000261680 UP000008225 UP000029965 UP000233020 UP000233080 UP000005640 UP000000589 UP000001519 UP000009130 UP000011712 UP000233040 UP000240080 UP000002277 UP000248482 UP000079721 UP000008912 UP000286641 UP000001075 UP000001073 UP000001646 UP000031443 UP000001811 UP000001595 UP000189706 UP000233160 UP000081671 UP000000715 UP000233180
UP000279307 UP000007151 UP000078492 UP000078542 UP000005203 UP000192223 UP000027135 UP000075809 UP000005205 UP000019118 UP000283509 UP000030742 UP000242457 UP000078541 UP000028761 UP000286642 UP000233120 UP000233100 UP000006718 UP000235965 UP000233140 UP000261680 UP000008225 UP000029965 UP000233020 UP000233080 UP000005640 UP000000589 UP000001519 UP000009130 UP000011712 UP000233040 UP000240080 UP000002277 UP000248482 UP000079721 UP000008912 UP000286641 UP000001075 UP000001073 UP000001646 UP000031443 UP000001811 UP000001595 UP000189706 UP000233160 UP000081671 UP000000715 UP000233180
Interpro
ProteinModelPortal
A0A2H1VZD8
A0A2A4J9M1
A0A194QR76
A0A194Q9E8
K7J5P2
D6WDB1
+ More
A0A154PPN4 A0A3L8DGV1 A0A212F819 A0A151IXH3 A0A195CY49 A0A087ZX87 A0A1B6E6L8 A0A224XJA2 A0A1W4XCH3 A0A069DQG6 A0A067R5L3 A0A151XH91 A0A158P2W8 N6TRD4 A0A3R7MJU5 A0A023FXU1 A0A1Y1M5B9 A0A0C9PRS7 U4U1W4 A0A2A3EI31 A0A2R5L5Q2 A0A195EZX8 A0A2I3LRI4 A0A3Q7X4X4 A0A2K6B163 A0A2K5U7C2 F7FQY3 A0A2J7QBD1 A0A2K5Z319 A0A384DT44 F6XBX7 A0A0D9S7N8 A0A2K5DYS2 A0A096MMW7 A0A2K5KC61 A0A2K5DYZ9 Q9H3H1-2 A0A2J8Y697 A0A3Q7XYH3 Q80UN9-2 A0A2K6B154 F7GJT8 I7G8M2 A0A2I2Z503 A0A2F0AWM5 H9F4Y3 G7NUC5 G7MG98 Q9H3H1-3 M3W562 A0A2K5RFZ3 A0A2K5Z325 A0A2K5DZ25 Q3T7C5 A0A2R8ZLW7 A0A2I3RL05 A0A2Y9J4F2 A0A1S3WGH9 G1LCG9 A0A3Q7P9Y3 G3HN83 D2HXL8 A0A2I3HS54 H9G668 A0A384DTX5 Q9H3H1-5 U3FSI0 M7BJR6 G1SGQ1 A0A1E1X3E8 A0A2K5KC58 H2N7V3 A0A1Z5L5F8 A0A2Y9IZV5 G3R3K0 A0A2K5DYN8 A0A2J8Y6C7 A0A3Q0DFI2 Q3T7C7 A0A293LL16 A0A2K5RFT6 A0A2K6EWL2 A0A1S3F8M3 A0A2R8ZPL5 H2PYR3 G1RN30 Q53F11 A0A1B6EHY5 Q9H3H1 A0A1Y1M5G9 M3YS73 A0A2K6MUL4
A0A154PPN4 A0A3L8DGV1 A0A212F819 A0A151IXH3 A0A195CY49 A0A087ZX87 A0A1B6E6L8 A0A224XJA2 A0A1W4XCH3 A0A069DQG6 A0A067R5L3 A0A151XH91 A0A158P2W8 N6TRD4 A0A3R7MJU5 A0A023FXU1 A0A1Y1M5B9 A0A0C9PRS7 U4U1W4 A0A2A3EI31 A0A2R5L5Q2 A0A195EZX8 A0A2I3LRI4 A0A3Q7X4X4 A0A2K6B163 A0A2K5U7C2 F7FQY3 A0A2J7QBD1 A0A2K5Z319 A0A384DT44 F6XBX7 A0A0D9S7N8 A0A2K5DYS2 A0A096MMW7 A0A2K5KC61 A0A2K5DYZ9 Q9H3H1-2 A0A2J8Y697 A0A3Q7XYH3 Q80UN9-2 A0A2K6B154 F7GJT8 I7G8M2 A0A2I2Z503 A0A2F0AWM5 H9F4Y3 G7NUC5 G7MG98 Q9H3H1-3 M3W562 A0A2K5RFZ3 A0A2K5Z325 A0A2K5DZ25 Q3T7C5 A0A2R8ZLW7 A0A2I3RL05 A0A2Y9J4F2 A0A1S3WGH9 G1LCG9 A0A3Q7P9Y3 G3HN83 D2HXL8 A0A2I3HS54 H9G668 A0A384DTX5 Q9H3H1-5 U3FSI0 M7BJR6 G1SGQ1 A0A1E1X3E8 A0A2K5KC58 H2N7V3 A0A1Z5L5F8 A0A2Y9IZV5 G3R3K0 A0A2K5DYN8 A0A2J8Y6C7 A0A3Q0DFI2 Q3T7C7 A0A293LL16 A0A2K5RFT6 A0A2K6EWL2 A0A1S3F8M3 A0A2R8ZPL5 H2PYR3 G1RN30 Q53F11 A0A1B6EHY5 Q9H3H1 A0A1Y1M5G9 M3YS73 A0A2K6MUL4
PDB
3EPL
E-value=9.17429e-18,
Score=218
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion
Cytoplasm
Nucleus
Cytoplasm
Nucleus
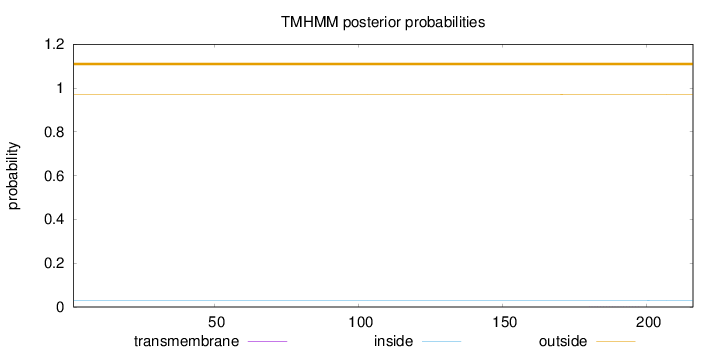
Length:
216
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00444
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02915
outside
1 - 216
Population Genetic Test Statistics
Pi
19.526511
Theta
17.340329
Tajima's D
0.327145
CLR
0.684099
CSRT
0.468576571171441
Interpretation
Uncertain
