Gene
KWMTBOMO06887
Annotation
reverse_transcriptase_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.426 PlasmaMembrane Reliability : 1.305
Sequence
CDS
ATGTGTTGTGCGACAAAACTAACTAAATCGCAGCCCGGCCCGGATGGTGTCCATGGCCGGGTTTGGGCCTTGGCTCTTGGTGCCCTAGGGGACCGACTCTTGAGACTTTACAACTCCTGCCTCGAGTCGGGACGGTTTCCTTCATCGTGGAGGACGGGCAGACTCGTGCTGTTGAGAAAGGAGGGGCGCCCGGCGGATACTGCCGCCGGGTACCGTCCCATCGTGTTGCTGGATGAGGTGGGCAAGCTGCTGGAACGCATTCTGGCAGCCCGCATCATCCAGCATCTAGTCGGGGTGGGACCCGATCTGTCGGCGGAGCAGTATGGCTTCCGAGAGGGCCGCTCAACGGTAGACGCGATCCTTCGCGTGCGGGCCCTCTCGGAGGAGGCCGTCTCCAGGGGTGGGGTGGCTTTGGCGGTGTCGCTCGATATCGCCAATGCGTTTAACACCCTGCCCTGGTCCGTGATAGGGGGGGCGCTGGAACGACATAGAGTGCCCCCCTACCTTCGCCGGCTGGTGGGTTCCTACTTAGAGGACAGATCGGTCACGTGTACCGGACACGGTGGGATCCTGCACCGGTTCCCGGTCGTGCGCGGTGTTCCACAGGGGTCGGTGCTCGGCCCCCTTTTGTGGAATATCGGGTATGACTGGGTGCTGAGAGGTGCCCTCCTCCCGGGCCTGAGCGTTATCTGTTACGCAGACGACACGTTGGTCGTGGCCCGGGGGGGAGTTTTGCTGAGTCTGCCCGTCTTGCTACGGCTGGGGCCCCGGAGAGTGCCACCTGTCGATGCCCATATCGTGGTTGGAGGCGTCCATATCGGGGTCGGGGTGCAGTTGAAGTACCTCGGCCTCATTCTGGACAGTCGTTGGACCTTCCGTGCTCACTTTCAGAACCTGGTCCCTCGTTTGTTGGGGGTGGCCGGCGCGTTAAGCCGGCTTCTGCCCAACGTCGGGGGGCCTGACCAGGTGACGCGCCGTCTCTATACGGGGGTGGTGCGATCAATGGCCCTATACGGGGCGCCCGTGTGGGGCCAGTCCCTGGCCGTGGGGGTGGCGAAGCTGCTGCAACGGCCGCAACGCACCATCGCGGTCAGGGTCATCCGTGGTTATCGCACCATCTCCTTTGAGGCGGCGTGTGTACTGGCTGGGACGCCGCCTTGGGTTCTGGAAGCGGAGGCGCTCGCCGCTGACTATAAGTGGCGGGCTGACCTTCGTGCCCGGGGCGTGGCGCGTCCCAGCCCCAGTGTGGTCAGAGCGCGGAGGGGCCAATCTCGGCGGTCCGTGCTGGAGTCATGGTCCAGACGGCTGGCCGATCCTTCGGCTGGTCGTAGGACCGTCGAGGCGATTCGCCCGGTCCTTGTGGATTGGGTGAATCGTGACAGAGGACGCCTCACTTTCCGGCTCACGCAGGTGCTCACTGGGCATGGTTGTTTCGGTGAGTTCCTGCACCGGATCGGAGCCGAGCCGACGGCAGAGTGCCACCATTGTGGTTGCGACTTGGACACGGCAGAGCATACGCTCGTCGCCTGCCCCGCATGGGAGGGGTGGCGCCGTGTCCTCGTCGCAAAAATAGGAACCGACTTGTCGTTGCCGAGTGTTGTGGCATCGATGCTCGGCGACGACGAGTCGTGGAAGGCGATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGCGTGAGAGACGCACAATCCCGCCGCCGTCGAGCGGGGGCCAGGGAGGCGGATCTCGCCCAAGCCCTGGCCCTCTAA
Protein
MCCATKLTKSQPGPDGVHGRVWALALGALGDRLLRLYNSCLESGRFPSSWRTGRLVLLRKEGRPADTAAGYRPIVLLDEVGKLLERILAARIIQHLVGVGPDLSAEQYGFREGRSTVDAILRVRALSEEAVSRGGVALAVSLDIANAFNTLPWSVIGGALERHRVPPYLRRLVGSYLEDRSVTCTGHGGILHRFPVVRGVPQGSVLGPLLWNIGYDWVLRGALLPGLSVICYADDTLVVARGGVLLSLPVLLRLGPRRVPPVDAHIVVGGVHIGVGVQLKYLGLILDSRWTFRAHFQNLVPRLLGVAGALSRLLPNVGGPDQVTRRLYTGVVRSMALYGAPVWGQSLAVGVAKLLQRPQRTIAVRVIRGYRTISFEAACVLAGTPPWVLEAEALAADYKWRADLRARGVARPSPSVVRARRGQSRRSVLESWSRRLADPSAGRRTVEAIRPVLVDWVNRDRGRLTFRLTQVLTGHGCFGEFLHRIGAEPTAECHHCGCDLDTAEHTLVACPAWEGWRRVLVAKIGTDLSLPSVVASMLGDDESWKAMLDFCECTISQKEAAGRVRDAQSRRRRAGAREADLAQALAL
Summary
Uniprot
Q868Q4
A0A2W1C3J5
A0A0J7KS67
A0A0J7N7S8
A0A0J7KLT9
A0A0J7KIY5
+ More
A0A2S2N8I2 A0A0J7KYD2 A0A0J7NEG9 A0A2S2PBP5 A0A0J7NAV8 A0A0J7JZ13 J9LJ80 A0A2S2PEZ7 A0A2S2NJ82 A0A2H8TIE5 J9JWJ3 A0A0J7KL06 A0A0J7MY17 A0A2S2P488 J9LVU7 X1WT13 A0A2S2PEU2 X1WMW4 A0A2S2NQQ6 J9KNG8 X1WXT5 J9KNN8 A0A142LX45 X1WSG8 A0A0J7KIS5 J9M6A5 J9M402 Q868S8 J9JZR8 J9KT61 Q868S0 A0A0J7K7P2 J9LB02 A0A034WR48 Q868S2 J9LQQ1 J9M8F6 A0A2H8THF8
A0A2S2N8I2 A0A0J7KYD2 A0A0J7NEG9 A0A2S2PBP5 A0A0J7NAV8 A0A0J7JZ13 J9LJ80 A0A2S2PEZ7 A0A2S2NJ82 A0A2H8TIE5 J9JWJ3 A0A0J7KL06 A0A0J7MY17 A0A2S2P488 J9LVU7 X1WT13 A0A2S2PEU2 X1WMW4 A0A2S2NQQ6 J9KNG8 X1WXT5 J9KNN8 A0A142LX45 X1WSG8 A0A0J7KIS5 J9M6A5 J9M402 Q868S8 J9JZR8 J9KT61 Q868S0 A0A0J7K7P2 J9LB02 A0A034WR48 Q868S2 J9LQQ1 J9M8F6 A0A2H8THF8
EMBL
AB090825
BAC57926.1
KZ149896
PZC78733.1
LBMM01003679
KMQ93242.1
+ More
LBMM01008682 KMQ88705.1 LBMM01005733 KMQ91252.1 LBMM01006954 KMQ90166.1 GGMR01000779 MBY13398.1 LBMM01002052 KMQ95336.1 LBMM01006110 KMQ90925.1 GGMR01014195 MBY26814.1 LBMM01007425 KMQ89740.1 LBMM01019839 KMQ83438.1 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 GGMR01015159 MBY27778.1 GGMR01004600 MBY17219.1 GFXV01002090 MBW13895.1 ABLF02013358 ABLF02013361 ABLF02054869 LBMM01006063 KMQ90967.1 LBMM01014122 KMQ85315.1 GGMR01011682 MBY24301.1 ABLF02034925 ABLF02014378 GGMR01015354 MBY27973.1 ABLF02059310 GGMR01006838 MBY19457.1 ABLF02005203 ABLF02041316 ABLF02004854 ABLF02041317 ABLF02023257 KU543683 AMS38371.1 ABLF02004597 ABLF02041324 LBMM01006977 KMQ90149.1 ABLF02020969 ABLF02028779 AB090813 BAC57902.1 ABLF02009709 ABLF02009711 ABLF02009073 ABLF02030702 ABLF02042963 AB090817 BAC57910.1 LBMM01012322 LBMM01012138 KMQ86282.1 KMQ86387.1 ABLF02011136 ABLF02014426 GAKP01002292 JAC56660.1 AB090816 BAC57908.1 ABLF02030663 ABLF02041312 ABLF02034556 GFXV01001748 MBW13553.1
LBMM01008682 KMQ88705.1 LBMM01005733 KMQ91252.1 LBMM01006954 KMQ90166.1 GGMR01000779 MBY13398.1 LBMM01002052 KMQ95336.1 LBMM01006110 KMQ90925.1 GGMR01014195 MBY26814.1 LBMM01007425 KMQ89740.1 LBMM01019839 KMQ83438.1 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 GGMR01015159 MBY27778.1 GGMR01004600 MBY17219.1 GFXV01002090 MBW13895.1 ABLF02013358 ABLF02013361 ABLF02054869 LBMM01006063 KMQ90967.1 LBMM01014122 KMQ85315.1 GGMR01011682 MBY24301.1 ABLF02034925 ABLF02014378 GGMR01015354 MBY27973.1 ABLF02059310 GGMR01006838 MBY19457.1 ABLF02005203 ABLF02041316 ABLF02004854 ABLF02041317 ABLF02023257 KU543683 AMS38371.1 ABLF02004597 ABLF02041324 LBMM01006977 KMQ90149.1 ABLF02020969 ABLF02028779 AB090813 BAC57902.1 ABLF02009709 ABLF02009711 ABLF02009073 ABLF02030702 ABLF02042963 AB090817 BAC57910.1 LBMM01012322 LBMM01012138 KMQ86282.1 KMQ86387.1 ABLF02011136 ABLF02014426 GAKP01002292 JAC56660.1 AB090816 BAC57908.1 ABLF02030663 ABLF02041312 ABLF02034556 GFXV01001748 MBW13553.1
Proteomes
PRIDE
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR002123 Plipid/glycerol_acylTrfase
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR026960 RVT-Znf
IPR021896 Transposase_37
IPR027806 HARBI1_dom
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR002123 Plipid/glycerol_acylTrfase
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR026960 RVT-Znf
IPR021896 Transposase_37
IPR027806 HARBI1_dom
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
Gene 3D
ProteinModelPortal
Q868Q4
A0A2W1C3J5
A0A0J7KS67
A0A0J7N7S8
A0A0J7KLT9
A0A0J7KIY5
+ More
A0A2S2N8I2 A0A0J7KYD2 A0A0J7NEG9 A0A2S2PBP5 A0A0J7NAV8 A0A0J7JZ13 J9LJ80 A0A2S2PEZ7 A0A2S2NJ82 A0A2H8TIE5 J9JWJ3 A0A0J7KL06 A0A0J7MY17 A0A2S2P488 J9LVU7 X1WT13 A0A2S2PEU2 X1WMW4 A0A2S2NQQ6 J9KNG8 X1WXT5 J9KNN8 A0A142LX45 X1WSG8 A0A0J7KIS5 J9M6A5 J9M402 Q868S8 J9JZR8 J9KT61 Q868S0 A0A0J7K7P2 J9LB02 A0A034WR48 Q868S2 J9LQQ1 J9M8F6 A0A2H8THF8
A0A2S2N8I2 A0A0J7KYD2 A0A0J7NEG9 A0A2S2PBP5 A0A0J7NAV8 A0A0J7JZ13 J9LJ80 A0A2S2PEZ7 A0A2S2NJ82 A0A2H8TIE5 J9JWJ3 A0A0J7KL06 A0A0J7MY17 A0A2S2P488 J9LVU7 X1WT13 A0A2S2PEU2 X1WMW4 A0A2S2NQQ6 J9KNG8 X1WXT5 J9KNN8 A0A142LX45 X1WSG8 A0A0J7KIS5 J9M6A5 J9M402 Q868S8 J9JZR8 J9KT61 Q868S0 A0A0J7K7P2 J9LB02 A0A034WR48 Q868S2 J9LQQ1 J9M8F6 A0A2H8THF8
PDB
6AR3
E-value=1.87459e-05,
Score=117
Ontologies
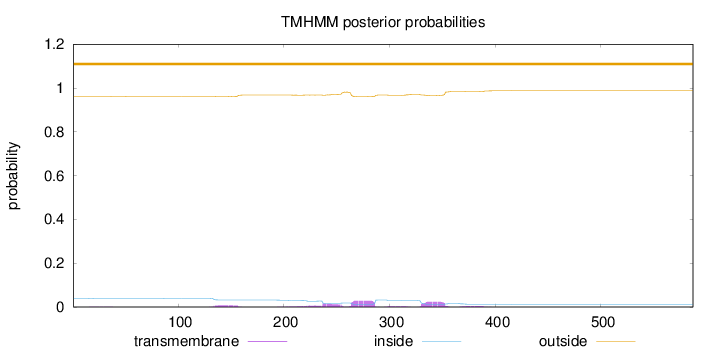
Topology
Length:
587
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.85851
Exp number, first 60 AAs:
0.00979
Total prob of N-in:
0.03971
outside
1 - 587
Population Genetic Test Statistics
Pi
1.231983
Theta
12.925709
Tajima's D
0
CLR
21.909504
CSRT
0.375831208439578
Interpretation
Uncertain
