Gene
KWMTBOMO06880 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012077
Annotation
PREDICTED:_lysosomal_alpha-glucosidase-like_isoform_X1_[Bombyx_mori]
Full name
Cytosolic Fe-S cluster assembly factor NUBP1 homolog
Location in the cell
Lysosomal Reliability : 0.976
Sequence
CDS
ATGCCTAAAATACCATATAAACCAGAAAAGTCTGGTGGAAATGACGATGACTACGAAATTGTTTCGTTCGATGATTTTTCCGACAAATCTCCGGGTTCGAAAACTGATTTATTAACTTTGAATGATAACATAAATTATCGTCTCTATTACGAAGCAGATAAACAATCAAAATATCTAGAAAACAATGGAGAATCATCCAAAGGTATTGACAGTCATACAGAAACGTCGCTGAATGGAGCAAGAAATGGATCATTCAGAAAATTATCAAATTCTTTTGTCCCATTCGGAAAATCCGGCGGGAAGACTCACACGCCGTTATTTAGCGGCGTTAAGTCTAAGAATGAAAATGAATCTTCATCTAGAGAACACAGATATCAACGATTTTCAGAAAATAGCACGTTTATGCAGAGAGTCTGCGATATGATGCCTGGAACTGGTTCAAGTGTACTAGCCATAATCTTGTTGTGCGGGTTGTCCCTTGCAAGTTATTGGGCAGTCGGCGGTACCATCGGTGGGTCCTGGGGCGAGGAGCATTACAAGAAACTATGGGAGAGAACGCATCCCGAAAATGTGAAGAAACCTCTATCACCCATGCCACATGACAAGCAGGTGATACCCGAATACAGGTACCACGACCACAACAACTTGAGCACCAAAAACAGAAGCAACGGAACTGAGGCTAAGAAGAAGGAATTCAGTAAGAAAAACGTGTACGTGGAACGTCCGGCCGCAGTCTTGCGCGAGCTATGCGCGCTCGTCGCGGACAATATGAAGTTCGACTGCTACCCTCAAGGCGACATCAATGAGGAGAACTGCGTCAAACGAGGTTGCTGCTGGAAGGTTACACAGATGAAGGGCGCTCCGTACTGCTACTACCCGTCGCAGTACGACACGTTCCGTTTCGTGAACATGACCGAGGACAAGCACTCCATGACCGTGTTCTACGAGAAGGCTCGACCCTCCGGTTATCCGGATGACTTTGAACTAGTCCGAATGGATTTTAAATACATGTCCGATGAAATTTTACAGATTAAGATATATGATGCGGAAAACAACCGATTCGAACCACCCTACCCAGAAATATCGATGGGCTCCAAGACCCTGACGGATATGAAATACAGAGTTGTCTTGGAAGGCTCGAAGATAGGGTTCAAAGTGATAAGGAATTCTGATGATGTTGTCATTTTCAACACCCAGGACGTGGGAGGGCTGATATTATCGGACAAGTTCCTTCAGATATCAGCAATACTTCCCACCGACCACGTGTTCGGCCTGGGAGAGAAGCGAGCGAGGTTCATGAACAACATGGACTGGCAGACCTTTGCCATATTCAACAGAGACAGGGCGCCCACGGAAGGGCTTAATTTGTATGGCACGCACCCTTTCTATCTGGCCGTGGAACAAAACGGACAAAGTCACGGGACGCTTCTACTCAATTCTAACGCTATGGACATAGTGCTGCAGCCGAGCCCCGGCATCACGTACCGCGCGACGGGCGGCGTGCTGCAGCTGTTCGTGCTGGCGGGCCCCAGCCCCGCGCTCGTCGCCCGCCAGTACACGGAGCTGGTGGGCAGGCCCTTCATGCCGCCCTACTGGTCCCTGGGCTTCCATCTGTGCAAGTTCAACTACGGGAACCTGAGCGTCATGGCGGACGTGTGGCAGAGGAACAGAGACGCTGGCATACCCTTTGACGTCCAGTGGAACGACATCGACTACATGACGCACAGCAACGACTTCACGCTGGACGCCGCCCGGTACGCGGGGCTGCCCGCTTTCGTGAAGAGGCTGCACTCCGAGGGCATGCACTACGTCATGATCTTTGATATTGGTATTGGAGCTTCAGAAAAGCCGGGCACGTATAAAGCCTACGACCGAGGCATAGAAATGGACATATTTGTGAAGAATTCTTCAAATCAACCGTTTATAGGAAGAGTCTGGAACAAAGGGTCCACGGTATATCCAGACTTCACTCATCCGAATTCGACTCCGTATTGGGTCGAAATGATGACGGACTTCCACAAACTCGTCCCGTACGACGGAGCGTGGATCGACATGAACGAGCCGTCGAACTTCGAGGACGGTCCCCTGCACGGCGCCTGCGCCCCCGAGCAGCTCCCGTACCGCCCGCGCACCGGCGACCCGCAGCTGCACACGCACACGCTGTGCATGGACGCCGCGCACCACGCCGGCCCGCACCGGCACTACCACGACCTGGTGGGGCTCATGCATGCCGTGGCCACCAACTTTGCCTTGCAAGAGATCAGAGGGAAGCGTCCGTTCATCATAAGCCGTGCGTCGTTCGTTGGACTGGGGAGGTACGCGGGCCACTGGAGCGGGGACATCAGCAGCCAGTGGCACGATATGAAGATGTCCATACCGGAGCTGCTGAGCTTCAGTCTGTTCGGAGTCCCCATGATGGGTGCCGACATCTGCGGCTTCAACGGCGACACCAACGTGGAGCTCTGCACGCGCTGGATGCAGCTCGGAGCCTTCTACCCCTTCTCGAGGAATCATAACACCGATACAGCCATTCCGCAAGACCCCGCGAGCCTGGGCCCGGCGGTGGTGCAGGCGAGCCGCGCCGCGCTGCGCACGCGCTACAAGCTGCTGCCCTACTACTACACGCTGTTCTGGCGCGCGCACGTGCTCGGCGACACCGTCGTGCGCCCGCTGCTCTTCGAATTTCCCGAGATGTCTGAGCTACATGACATCGACACCCAGTTCATGGTCGGGCCGCACGTGATGGTCGCGCCGGTGCTGGAGCCCGGCACGGACCGCGCCCGCGCCCTGTTCCCCGGCGCGCAGGGCTGGTACAGCCTGGCGACGGGCCGACACATCGCCACCAACGATTGGGCCGAAGTCTTGCACAACGACACTGTCTCTATTAAGGGCGGCGGCGTGGTGGTGCTGCAGGAGCCGGCGGCGCGCGGGCCCGTCAGCACCGGCTCGGCCCGCAGCGCGCCGCTGCAGCTGCTCGCGGCGCCCGACGCAGCCGCCGCCGCAAGCGGACGCCTCTACTGGGACGACGGCGACAGCCTGCACACCTACGAAGAGAAGAAATACAGTTACATAGAATTTTCACTGAACGGCACTGAAATCCAATCACATGTGAAATGGTGGGGATATGGAGTTCCGCTGTTCAACCAGCTGTCAATTCTCAATCAGCCCCCCGTCAAGCAAGTTCTGATCAACGGAATGCCGTGCGTGTCCCCCTGCTCGTTCACGTACTACAAGAAGAACAAAGTGTTGTCCATTATGATTAATTTGTCATTAGAGAAACCATTCAACGTCAGATGGTCATACGATGAGAAATATTTGAACAGGACCGGGGCATCGAGAAAAAATTGA
Protein
MPKIPYKPEKSGGNDDDYEIVSFDDFSDKSPGSKTDLLTLNDNINYRLYYEADKQSKYLENNGESSKGIDSHTETSLNGARNGSFRKLSNSFVPFGKSGGKTHTPLFSGVKSKNENESSSREHRYQRFSENSTFMQRVCDMMPGTGSSVLAIILLCGLSLASYWAVGGTIGGSWGEEHYKKLWERTHPENVKKPLSPMPHDKQVIPEYRYHDHNNLSTKNRSNGTEAKKKEFSKKNVYVERPAAVLRELCALVADNMKFDCYPQGDINEENCVKRGCCWKVTQMKGAPYCYYPSQYDTFRFVNMTEDKHSMTVFYEKARPSGYPDDFELVRMDFKYMSDEILQIKIYDAENNRFEPPYPEISMGSKTLTDMKYRVVLEGSKIGFKVIRNSDDVVIFNTQDVGGLILSDKFLQISAILPTDHVFGLGEKRARFMNNMDWQTFAIFNRDRAPTEGLNLYGTHPFYLAVEQNGQSHGTLLLNSNAMDIVLQPSPGITYRATGGVLQLFVLAGPSPALVARQYTELVGRPFMPPYWSLGFHLCKFNYGNLSVMADVWQRNRDAGIPFDVQWNDIDYMTHSNDFTLDAARYAGLPAFVKRLHSEGMHYVMIFDIGIGASEKPGTYKAYDRGIEMDIFVKNSSNQPFIGRVWNKGSTVYPDFTHPNSTPYWVEMMTDFHKLVPYDGAWIDMNEPSNFEDGPLHGACAPEQLPYRPRTGDPQLHTHTLCMDAAHHAGPHRHYHDLVGLMHAVATNFALQEIRGKRPFIISRASFVGLGRYAGHWSGDISSQWHDMKMSIPELLSFSLFGVPMMGADICGFNGDTNVELCTRWMQLGAFYPFSRNHNTDTAIPQDPASLGPAVVQASRAALRTRYKLLPYYYTLFWRAHVLGDTVVRPLLFEFPEMSELHDIDTQFMVGPHVMVAPVLEPGTDRARALFPGAQGWYSLATGRHIATNDWAEVLHNDTVSIKGGGVVVLQEPAARGPVSTGSARSAPLQLLAAPDAAAAASGRLYWDDGDSLHTYEEKKYSYIEFSLNGTEIQSHVKWWGYGVPLFNQLSILNQPPVKQVLINGMPCVSPCSFTYYKKNKVLSIMINLSLEKPFNVRWSYDEKYLNRTGASRKN
Summary
Description
Component of the cytosolic iron-sulfur (Fe/S) protein assembly (CIA) machinery. Required for maturation of extramitochondrial Fe-S proteins. The NUBP1-NUBP2 heterotetramer forms a Fe-S scaffold complex, mediating the de novo assembly of an Fe-S cluster and its transfer to target apoproteins.
Cofactor
[4Fe-4S] cluster
Subunit
Heterotetramer of 2 NUBP1 and 2 NUBP2 chains.
Similarity
Belongs to the glycosyl hydrolase 31 family.
Belongs to the Mrp/NBP35 ATP-binding proteins family. NUBP1/NBP35 subfamily.
Belongs to the Mrp/NBP35 ATP-binding proteins family. NUBP1/NBP35 subfamily.
Uniprot
H9JRB7
A0A2A4K069
A0A3S2NXS9
A0A212F808
A0A194QAX6
A0A194QQ36
+ More
A0A067R6F4 D6WDN8 A0A1Y1L9Q9 A0A1W4X0K2 A0A1W4WPS6 A0A1W4X0S9 A0A1W4WPS7 A0A1W4WZI1 N6UEN5 U4URX8 A0A154PP69 A0A1B6F1T2 E2BEF1 A0A232EPJ9 A0A088AP40 E9INW4 A0A195CTJ7 E9H648 E2A599 A0A0L7R0H2 A0A2H8TF00 A0A2A3E141 A0A0J7L2F7 A0A0P5WPA1 A0A0P5WQY7 A0A151WZI2 A0A0P5XJE2 A0A0N8CYY9 A0A158NT86 A0A0P5WK62 A0A0P5VR91 A0A0P5Y9E2 A0A0P5SLI1 A0A0P5YQP4 A0A0P5VMU5 A0A0P5EBP1 A0A0P6A4I4 A0A0P5BFG8 A0A0P5UZ20 A0A0P5SCL6 A0A0P5VQY0 A0A0N8CE76 A0A0P5TGZ7 A0A0P5XVY9 A0A0N8D313 A0A0P5TQQ9 A0A0P5SYE4 A0A0P5ECX5 A0A0N8E534 A0A0P5VH76 A0A0P5EKD1 A0A0P5AQD2 A0A0P5HER8 A0A0P5KSM1 A0A0N7ZS03 A0A0N7ZSQ9 A0A195E332 A0A1S6J0W8 A0A0P5V4A1 A0A195BSZ9 J9JP90 A0A0N8A1L5 A0A151JVQ8 A0A0P5CX84 A0A0P5KGJ0 A0A0P5PRM3 A0A0N8A9J0 A0A0P5K557 A0A0P5NCV0 K7IPK3 F4WH36 A0A0P5KBP7 A0A0N8E9A8 A0A0P6HR96 A0A0P6GG39 A0A0N8B239 A0A0P5QVL2 A0A0P5QH45 A0A1B6EMJ2 A0A0N8AEN9 A0A0P5WH59 A0A0P6AQT6 A0A0P5VMH1 A0A0P6B2W6 A0A0P4YCU2 A0A3L8D368 A0A1B6D5E9 A0A0P4W3H6
A0A067R6F4 D6WDN8 A0A1Y1L9Q9 A0A1W4X0K2 A0A1W4WPS6 A0A1W4X0S9 A0A1W4WPS7 A0A1W4WZI1 N6UEN5 U4URX8 A0A154PP69 A0A1B6F1T2 E2BEF1 A0A232EPJ9 A0A088AP40 E9INW4 A0A195CTJ7 E9H648 E2A599 A0A0L7R0H2 A0A2H8TF00 A0A2A3E141 A0A0J7L2F7 A0A0P5WPA1 A0A0P5WQY7 A0A151WZI2 A0A0P5XJE2 A0A0N8CYY9 A0A158NT86 A0A0P5WK62 A0A0P5VR91 A0A0P5Y9E2 A0A0P5SLI1 A0A0P5YQP4 A0A0P5VMU5 A0A0P5EBP1 A0A0P6A4I4 A0A0P5BFG8 A0A0P5UZ20 A0A0P5SCL6 A0A0P5VQY0 A0A0N8CE76 A0A0P5TGZ7 A0A0P5XVY9 A0A0N8D313 A0A0P5TQQ9 A0A0P5SYE4 A0A0P5ECX5 A0A0N8E534 A0A0P5VH76 A0A0P5EKD1 A0A0P5AQD2 A0A0P5HER8 A0A0P5KSM1 A0A0N7ZS03 A0A0N7ZSQ9 A0A195E332 A0A1S6J0W8 A0A0P5V4A1 A0A195BSZ9 J9JP90 A0A0N8A1L5 A0A151JVQ8 A0A0P5CX84 A0A0P5KGJ0 A0A0P5PRM3 A0A0N8A9J0 A0A0P5K557 A0A0P5NCV0 K7IPK3 F4WH36 A0A0P5KBP7 A0A0N8E9A8 A0A0P6HR96 A0A0P6GG39 A0A0N8B239 A0A0P5QVL2 A0A0P5QH45 A0A1B6EMJ2 A0A0N8AEN9 A0A0P5WH59 A0A0P6AQT6 A0A0P5VMH1 A0A0P6B2W6 A0A0P4YCU2 A0A3L8D368 A0A1B6D5E9 A0A0P4W3H6
Pubmed
EMBL
BABH01016069
BABH01016070
BABH01016071
NWSH01000295
PCG77661.1
RSAL01000107
+ More
RVE47283.1 AGBW02009812 OWR49877.1 KQ459252 KPJ02145.1 KQ461185 KPJ07474.1 KK852855 KDR14932.1 KQ971322 EEZ99945.2 GEZM01061536 GEZM01061535 JAV70402.1 APGK01038392 KB740954 ENN77102.1 KB632338 ERL92906.1 KQ435007 KZC13685.1 GECZ01025718 JAS44051.1 GL447768 EFN85996.1 NNAY01002918 OXU20309.1 GL764397 EFZ17773.1 KQ977329 KYN03454.1 GL732596 EFX72744.1 GL436894 EFN71395.1 KQ414670 KOC64342.1 GFXV01000854 MBW12659.1 KZ288483 PBC25244.1 LBMM01001148 KMQ96823.1 GDIP01083482 JAM20233.1 GDIP01083481 JAM20234.1 KQ982636 KYQ53310.1 GDIP01083483 JAM20232.1 GDIP01085135 JAM18580.1 ADTU01025716 GDIP01085134 JAM18581.1 GDIP01096642 JAM07073.1 GDIP01061188 JAM42527.1 GDIP01138273 JAL65441.1 GDIP01055037 JAM48678.1 GDIP01097885 JAM05830.1 GDIP01143678 JAJ79724.1 GDIP01047332 JAM56383.1 GDIP01200426 JAJ22976.1 GDIP01109532 JAL94182.1 GDIP01141480 GDIP01140440 JAL62234.1 GDIP01113691 GDIP01096643 JAM07072.1 GDIP01140439 JAL63275.1 GDIP01126376 JAL77338.1 GDIP01231777 GDIP01218312 GDIP01209165 GDIP01200425 GDIP01199064 GDIP01139380 GDIP01133237 GDIP01131987 GDIP01130632 GDIP01125223 GDIP01122927 GDIP01086434 GDIP01066637 GDIP01063940 GDIP01050894 GDIP01045944 GDIP01044448 JAM37078.1 GDIP01240574 GDIP01239481 GDIP01219724 GDIP01198027 GDIP01196017 GDIP01160969 GDIP01157071 GDIP01156027 GDIP01124220 GDIP01109530 GDIP01094379 GDIP01089736 GDIP01087533 GDIP01073743 GDIP01072636 GDIP01071336 GDIP01052155 GDIQ01067196 JAM29972.1 JAN27541.1 GDIP01122928 JAL80786.1 GDIP01138274 JAL65440.1 GDIP01143679 JAJ79723.1 GDIQ01060996 JAN33741.1 GDIP01115035 JAL88679.1 GDIP01240575 GDIP01239482 GDIP01220876 GDIP01219723 GDIP01198028 GDIP01165672 GDIP01164972 GDIP01158579 GDIP01157070 GDIP01156026 GDIP01145730 GDIQ01167853 GDIQ01152507 GDIP01124219 GDIP01109531 GDIP01094378 GDIP01087532 GDIP01073744 GDIP01072637 GDIP01071337 GDIP01052156 GDIQ01067245 LRGB01000642 JAJ64823.1 JAK83872.1 KZS17340.1 GDIP01230519 GDIP01200424 GDIP01199063 GDIP01197178 GDIP01167170 GDIP01158024 GDIP01139381 GDIP01133238 GDIP01130631 GDIP01125222 GDIP01122926 GDIP01086435 GDIP01063941 GDIP01044447 GDIP01040391 JAJ22978.1 GDIQ01230834 JAK20891.1 GDIQ01179790 JAK71935.1 GDIP01218313 JAJ05089.1 GDIP01216265 JAJ07137.1 KQ979701 KYN19575.1 KU764423 AQS60670.1 GDIP01104626 JAL99088.1 KQ976409 KYM90922.1 ABLF02032902 GDIP01191417 JAJ31985.1 KQ981686 KYN37953.1 GDIP01164171 JAJ59231.1 GDIQ01186042 JAK65683.1 GDIQ01145461 JAL06265.1 GDIP01169217 JAJ54185.1 GDIQ01189102 JAK62623.1 GDIQ01147069 JAL04657.1 GL888148 EGI66515.1 GDIQ01187649 JAK64076.1 GDIQ01049204 JAN45533.1 GDIQ01016544 JAN78193.1 GDIQ01053707 JAN41030.1 GDIQ01220347 JAK31378.1 GDIQ01129314 JAL22412.1 GDIQ01117236 JAL34490.1 GECZ01030630 GECZ01023675 JAS39139.1 JAS46094.1 GDIP01154825 JAJ68577.1 GDIP01099273 JAM04442.1 GDIP01039597 JAM64118.1 GDIP01099274 JAM04441.1 GDIP01021205 JAM82510.1 GDIP01232918 JAI90483.1 QOIP01000014 RLU14947.1 GEDC01016377 JAS20921.1 GDRN01075433 JAI63060.1
RVE47283.1 AGBW02009812 OWR49877.1 KQ459252 KPJ02145.1 KQ461185 KPJ07474.1 KK852855 KDR14932.1 KQ971322 EEZ99945.2 GEZM01061536 GEZM01061535 JAV70402.1 APGK01038392 KB740954 ENN77102.1 KB632338 ERL92906.1 KQ435007 KZC13685.1 GECZ01025718 JAS44051.1 GL447768 EFN85996.1 NNAY01002918 OXU20309.1 GL764397 EFZ17773.1 KQ977329 KYN03454.1 GL732596 EFX72744.1 GL436894 EFN71395.1 KQ414670 KOC64342.1 GFXV01000854 MBW12659.1 KZ288483 PBC25244.1 LBMM01001148 KMQ96823.1 GDIP01083482 JAM20233.1 GDIP01083481 JAM20234.1 KQ982636 KYQ53310.1 GDIP01083483 JAM20232.1 GDIP01085135 JAM18580.1 ADTU01025716 GDIP01085134 JAM18581.1 GDIP01096642 JAM07073.1 GDIP01061188 JAM42527.1 GDIP01138273 JAL65441.1 GDIP01055037 JAM48678.1 GDIP01097885 JAM05830.1 GDIP01143678 JAJ79724.1 GDIP01047332 JAM56383.1 GDIP01200426 JAJ22976.1 GDIP01109532 JAL94182.1 GDIP01141480 GDIP01140440 JAL62234.1 GDIP01113691 GDIP01096643 JAM07072.1 GDIP01140439 JAL63275.1 GDIP01126376 JAL77338.1 GDIP01231777 GDIP01218312 GDIP01209165 GDIP01200425 GDIP01199064 GDIP01139380 GDIP01133237 GDIP01131987 GDIP01130632 GDIP01125223 GDIP01122927 GDIP01086434 GDIP01066637 GDIP01063940 GDIP01050894 GDIP01045944 GDIP01044448 JAM37078.1 GDIP01240574 GDIP01239481 GDIP01219724 GDIP01198027 GDIP01196017 GDIP01160969 GDIP01157071 GDIP01156027 GDIP01124220 GDIP01109530 GDIP01094379 GDIP01089736 GDIP01087533 GDIP01073743 GDIP01072636 GDIP01071336 GDIP01052155 GDIQ01067196 JAM29972.1 JAN27541.1 GDIP01122928 JAL80786.1 GDIP01138274 JAL65440.1 GDIP01143679 JAJ79723.1 GDIQ01060996 JAN33741.1 GDIP01115035 JAL88679.1 GDIP01240575 GDIP01239482 GDIP01220876 GDIP01219723 GDIP01198028 GDIP01165672 GDIP01164972 GDIP01158579 GDIP01157070 GDIP01156026 GDIP01145730 GDIQ01167853 GDIQ01152507 GDIP01124219 GDIP01109531 GDIP01094378 GDIP01087532 GDIP01073744 GDIP01072637 GDIP01071337 GDIP01052156 GDIQ01067245 LRGB01000642 JAJ64823.1 JAK83872.1 KZS17340.1 GDIP01230519 GDIP01200424 GDIP01199063 GDIP01197178 GDIP01167170 GDIP01158024 GDIP01139381 GDIP01133238 GDIP01130631 GDIP01125222 GDIP01122926 GDIP01086435 GDIP01063941 GDIP01044447 GDIP01040391 JAJ22978.1 GDIQ01230834 JAK20891.1 GDIQ01179790 JAK71935.1 GDIP01218313 JAJ05089.1 GDIP01216265 JAJ07137.1 KQ979701 KYN19575.1 KU764423 AQS60670.1 GDIP01104626 JAL99088.1 KQ976409 KYM90922.1 ABLF02032902 GDIP01191417 JAJ31985.1 KQ981686 KYN37953.1 GDIP01164171 JAJ59231.1 GDIQ01186042 JAK65683.1 GDIQ01145461 JAL06265.1 GDIP01169217 JAJ54185.1 GDIQ01189102 JAK62623.1 GDIQ01147069 JAL04657.1 GL888148 EGI66515.1 GDIQ01187649 JAK64076.1 GDIQ01049204 JAN45533.1 GDIQ01016544 JAN78193.1 GDIQ01053707 JAN41030.1 GDIQ01220347 JAK31378.1 GDIQ01129314 JAL22412.1 GDIQ01117236 JAL34490.1 GECZ01030630 GECZ01023675 JAS39139.1 JAS46094.1 GDIP01154825 JAJ68577.1 GDIP01099273 JAM04442.1 GDIP01039597 JAM64118.1 GDIP01099274 JAM04441.1 GDIP01021205 JAM82510.1 GDIP01232918 JAI90483.1 QOIP01000014 RLU14947.1 GEDC01016377 JAS20921.1 GDRN01075433 JAI63060.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000027135 UP000007266 UP000192223 UP000019118 UP000030742 UP000076502 UP000008237 UP000215335 UP000005203 UP000078542 UP000000305 UP000000311 UP000053825 UP000242457 UP000036403 UP000075809 UP000005205 UP000076858 UP000078492 UP000078540 UP000007819 UP000078541 UP000002358 UP000007755 UP000279307
UP000027135 UP000007266 UP000192223 UP000019118 UP000030742 UP000076502 UP000008237 UP000215335 UP000005203 UP000078542 UP000000305 UP000000311 UP000053825 UP000242457 UP000036403 UP000075809 UP000005205 UP000076858 UP000078492 UP000078540 UP000007819 UP000078541 UP000002358 UP000007755 UP000279307
PRIDE
Pfam
Interpro
IPR030459
Glyco_hydro_31_CS
+ More
IPR011013 Gal_mutarotase_sf_dom
IPR000322 Glyco_hydro_31
IPR025887 Glyco_hydro_31_N_dom
IPR017853 Glycoside_hydrolase_SF
IPR000519 P_trefoil_dom
IPR031727 Gal_mutarotase_N
IPR013780 Glyco_hydro_b
IPR030458 Glyco_hydro_31_AS
IPR017957 P_trefoil_CS
IPR033403 DUF5110
IPR033756 YlxH/NBP35
IPR000808 Mrp_CS
IPR028601 NUBP1/Nbp35
IPR027417 P-loop_NTPase
IPR019591 Mrp/NBP35_ATP-bd
IPR011013 Gal_mutarotase_sf_dom
IPR000322 Glyco_hydro_31
IPR025887 Glyco_hydro_31_N_dom
IPR017853 Glycoside_hydrolase_SF
IPR000519 P_trefoil_dom
IPR031727 Gal_mutarotase_N
IPR013780 Glyco_hydro_b
IPR030458 Glyco_hydro_31_AS
IPR017957 P_trefoil_CS
IPR033403 DUF5110
IPR033756 YlxH/NBP35
IPR000808 Mrp_CS
IPR028601 NUBP1/Nbp35
IPR027417 P-loop_NTPase
IPR019591 Mrp/NBP35_ATP-bd
Gene 3D
ProteinModelPortal
H9JRB7
A0A2A4K069
A0A3S2NXS9
A0A212F808
A0A194QAX6
A0A194QQ36
+ More
A0A067R6F4 D6WDN8 A0A1Y1L9Q9 A0A1W4X0K2 A0A1W4WPS6 A0A1W4X0S9 A0A1W4WPS7 A0A1W4WZI1 N6UEN5 U4URX8 A0A154PP69 A0A1B6F1T2 E2BEF1 A0A232EPJ9 A0A088AP40 E9INW4 A0A195CTJ7 E9H648 E2A599 A0A0L7R0H2 A0A2H8TF00 A0A2A3E141 A0A0J7L2F7 A0A0P5WPA1 A0A0P5WQY7 A0A151WZI2 A0A0P5XJE2 A0A0N8CYY9 A0A158NT86 A0A0P5WK62 A0A0P5VR91 A0A0P5Y9E2 A0A0P5SLI1 A0A0P5YQP4 A0A0P5VMU5 A0A0P5EBP1 A0A0P6A4I4 A0A0P5BFG8 A0A0P5UZ20 A0A0P5SCL6 A0A0P5VQY0 A0A0N8CE76 A0A0P5TGZ7 A0A0P5XVY9 A0A0N8D313 A0A0P5TQQ9 A0A0P5SYE4 A0A0P5ECX5 A0A0N8E534 A0A0P5VH76 A0A0P5EKD1 A0A0P5AQD2 A0A0P5HER8 A0A0P5KSM1 A0A0N7ZS03 A0A0N7ZSQ9 A0A195E332 A0A1S6J0W8 A0A0P5V4A1 A0A195BSZ9 J9JP90 A0A0N8A1L5 A0A151JVQ8 A0A0P5CX84 A0A0P5KGJ0 A0A0P5PRM3 A0A0N8A9J0 A0A0P5K557 A0A0P5NCV0 K7IPK3 F4WH36 A0A0P5KBP7 A0A0N8E9A8 A0A0P6HR96 A0A0P6GG39 A0A0N8B239 A0A0P5QVL2 A0A0P5QH45 A0A1B6EMJ2 A0A0N8AEN9 A0A0P5WH59 A0A0P6AQT6 A0A0P5VMH1 A0A0P6B2W6 A0A0P4YCU2 A0A3L8D368 A0A1B6D5E9 A0A0P4W3H6
A0A067R6F4 D6WDN8 A0A1Y1L9Q9 A0A1W4X0K2 A0A1W4WPS6 A0A1W4X0S9 A0A1W4WPS7 A0A1W4WZI1 N6UEN5 U4URX8 A0A154PP69 A0A1B6F1T2 E2BEF1 A0A232EPJ9 A0A088AP40 E9INW4 A0A195CTJ7 E9H648 E2A599 A0A0L7R0H2 A0A2H8TF00 A0A2A3E141 A0A0J7L2F7 A0A0P5WPA1 A0A0P5WQY7 A0A151WZI2 A0A0P5XJE2 A0A0N8CYY9 A0A158NT86 A0A0P5WK62 A0A0P5VR91 A0A0P5Y9E2 A0A0P5SLI1 A0A0P5YQP4 A0A0P5VMU5 A0A0P5EBP1 A0A0P6A4I4 A0A0P5BFG8 A0A0P5UZ20 A0A0P5SCL6 A0A0P5VQY0 A0A0N8CE76 A0A0P5TGZ7 A0A0P5XVY9 A0A0N8D313 A0A0P5TQQ9 A0A0P5SYE4 A0A0P5ECX5 A0A0N8E534 A0A0P5VH76 A0A0P5EKD1 A0A0P5AQD2 A0A0P5HER8 A0A0P5KSM1 A0A0N7ZS03 A0A0N7ZSQ9 A0A195E332 A0A1S6J0W8 A0A0P5V4A1 A0A195BSZ9 J9JP90 A0A0N8A1L5 A0A151JVQ8 A0A0P5CX84 A0A0P5KGJ0 A0A0P5PRM3 A0A0N8A9J0 A0A0P5K557 A0A0P5NCV0 K7IPK3 F4WH36 A0A0P5KBP7 A0A0N8E9A8 A0A0P6HR96 A0A0P6GG39 A0A0N8B239 A0A0P5QVL2 A0A0P5QH45 A0A1B6EMJ2 A0A0N8AEN9 A0A0P5WH59 A0A0P6AQT6 A0A0P5VMH1 A0A0P6B2W6 A0A0P4YCU2 A0A3L8D368 A0A1B6D5E9 A0A0P4W3H6
PDB
5NN8
E-value=0,
Score=1686
Ontologies
PATHWAY
GO
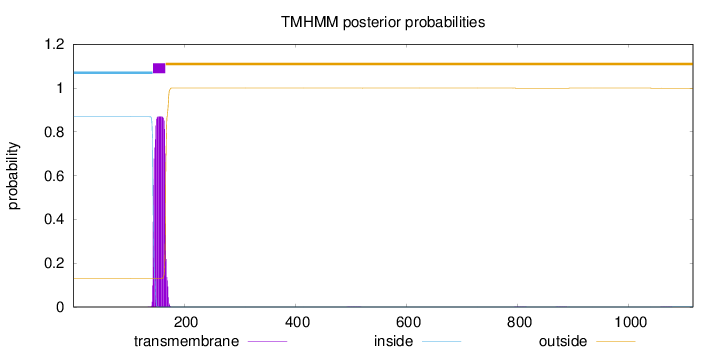
Topology
Subcellular location
Cytoplasm
Length:
1115
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.6402699999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.86889
inside
1 - 143
TMhelix
144 - 166
outside
167 - 1115
Population Genetic Test Statistics
Pi
16.441109
Theta
16.881454
Tajima's D
-0.923188
CLR
1.311788
CSRT
0.150042497875106
Interpretation
Uncertain
