Gene
KWMTBOMO06876
Pre Gene Modal
BGIBMGA012078
Annotation
PREDICTED:_sorting_nexin-14-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.44 Mitochondrial Reliability : 1.291 Nuclear Reliability : 1.643
Sequence
CDS
ATGAAGATCTTAAATCCGGATTTGTCTGAAAGCGAGAAGGCTTCATTGCACGGCGAGGCCTGCGACATCTACTCCGTGTACCTGTCGCAACGCTCCGCCCACAGAGTGCCGCTGCACCCCGCGCTGGCCACGGAGCTGTTCGAGCTGCTGCAGAGCCGGGACAGCATCAGGAGACTGCAAACAAGCCGCGCCCTGTACCAAGCGGCCAGACAATCGCACGCCGTACTCGAAAAGATAATGTTGCCGAGATTCCTGCACAGCGAACAGTTTTATAAATTGTTATTAGGAAGCCGAATACCCACCGGATATCAGAAGCACATATTCAAGAAGCCCCAGGAGAAGCTTCTGAACACCGCCATAAGGCTGGGCGGCAGACTGCGCGGCGCCCTCAGGGCACCCGCCGCCGACGGGCAGCTGCCGGACGGCTACCCCGACTCGGATGAGCTGGAGGCCAACGGCGACGACAACATCGACATCCTCAAGTATCTGGAGAGCATCACGGCCGAGGAGTCTCTGCGGGGGAACGACCTGACCAAGTACAAGGTCGTGCTCACCAATGTCGAGATCGTGCAGGCGCCCCCGCGGCGCGGCGGCGTGCGCGCGTTCACGCTGGGCGTGCAGCGGCTGGAGGGCGCGCTGTGCTACCGCGTGCTGCGCCGCACGGAGCACGACTTCCACCTGCTGCGGAGCAAGCTGCACGAGTTCCACGGGGACGCGCTGCTGCACGACCTGCCGCTGCCCTCCAGGAGAGACAACAGTCCATTGGAGACTATACGGTACAAGTACGAAGATTTCCTTCAGCGTCTGCTGCAGAAGAGTCTGTTGCAGACGAGCGAGCTGCTCAGGCTGTTCCTCACGGAGGAGGGAGACTTTTCCCTGGCGGTCCAGGCGTCGACCCTCAACGCAAGCAGCACAGACCTGCTGAACATCTATCAGTCCGTCACCCACAAGCTGAGGAAGGAGAAGGGGCAGCACCTGGAGAGCTTCCTGAGGAATCTGCTCGTGTCTTCAGACAGGGACCGGTACCGGGCACTCAAACAAGGTCGAGAGGTGGAGGAGGCGGTGGAGGTCAGCGAAGAGGTGGAGGCGGAGCAGCGGAGCGCCGGGCCGCGGCGCCCGCGCGACCTCACGGCCTCCGTGTTTGGGAACAACTTCGACGTGGAGTCCGGCCCCGTTACGTCAGCGCCCGCCCACCGGACCCCCGTCGCCGGCCTCACTCAGTGCCTCATGTATCTGTTGTGCGGGGCGGGGGGCGCGCGCGGGGTGGCGGCGGGCGCGCTGGGGGCGGCGCTGGGCTGCGTGCGCGCGCCGCTGGACGCCGCCGCCGACGCGCACCTCGCCGCCGCGCTGCGCCGCACGCTCACCCAGCCGCGCCTGGCGCATCTCATCGCGCTGGGACAAGAATTATTATTCGGTAAGAGATCTTCAACTCCACGTGTAGACTCGAACGAGCATCGTGATCTGGCTCGGCGACAGTTGCTGGGCGTTACGCGGGGCGCCGCGATGCTGGTCCCAGGACTAAAGGACGCGCTCAACAACGCCTTCGAGATTATACAGGACCCGAGGCTGAATAAACAGTTGGTGTATAATTTATTGGACCTGTGTTTAGTGGAGCTGTTTCCGGAATTGAAAAACGAACATGAATGTTCCGACAGCGGACCAAAGAGCTGA
Protein
MKILNPDLSESEKASLHGEACDIYSVYLSQRSAHRVPLHPALATELFELLQSRDSIRRLQTSRALYQAARQSHAVLEKIMLPRFLHSEQFYKLLLGSRIPTGYQKHIFKKPQEKLLNTAIRLGGRLRGALRAPAADGQLPDGYPDSDELEANGDDNIDILKYLESITAEESLRGNDLTKYKVVLTNVEIVQAPPRRGGVRAFTLGVQRLEGALCYRVLRRTEHDFHLLRSKLHEFHGDALLHDLPLPSRRDNSPLETIRYKYEDFLQRLLQKSLLQTSELLRLFLTEEGDFSLAVQASTLNASSTDLLNIYQSVTHKLRKEKGQHLESFLRNLLVSSDRDRYRALKQGREVEEAVEVSEEVEAEQRSAGPRRPRDLTASVFGNNFDVESGPVTSAPAHRTPVAGLTQCLMYLLCGAGGARGVAAGALGAALGCVRAPLDAAADAHLAAALRRTLTQPRLAHLIALGQELLFGKRSSTPRVDSNEHRDLARRQLLGVTRGAAMLVPGLKDALNNAFEIIQDPRLNKQLVYNLLDLCLVELFPELKNEHECSDSGPKS
Summary
Uniprot
H9JRB8
A0A2H1V4G1
A0A3S2PKW8
A0A2A4K067
A0A1E1W5V1
A0A2A4K195
+ More
A0A194Q9F6 A0A2A4K0A2 A0A212EYM0 D6WEY6 A0A1Y1LTS8 A0A2J7QAL2 A0A2J7QAI0 A0A067R2I0 A0A2J7QAI1 E2B9W5 E2AGL5 A0A195CLP3 A0A151J8S2 A0A088AJL1 A0A158P1S7 A0A0L7RE78 A0A151XAX9 F4W6N9 A0A1W4WR76 A0A0M8ZMN9 A0A2P8XKS6 A0A1B6EFM1 A0A1B6CSG8 T1HCS5 A0A1B6FTB1 A0A224XJT2 R7TFG3 A0A069DX22 A0A0V0G5S2 C3Z7E0 A0A1B6HIN5 A0A1B6KT93 A0A1B6JRI3 A0A0K8T7K7 A0A3Q0IRY1 A0A1B6M0E3
A0A194Q9F6 A0A2A4K0A2 A0A212EYM0 D6WEY6 A0A1Y1LTS8 A0A2J7QAL2 A0A2J7QAI0 A0A067R2I0 A0A2J7QAI1 E2B9W5 E2AGL5 A0A195CLP3 A0A151J8S2 A0A088AJL1 A0A158P1S7 A0A0L7RE78 A0A151XAX9 F4W6N9 A0A1W4WR76 A0A0M8ZMN9 A0A2P8XKS6 A0A1B6EFM1 A0A1B6CSG8 T1HCS5 A0A1B6FTB1 A0A224XJT2 R7TFG3 A0A069DX22 A0A0V0G5S2 C3Z7E0 A0A1B6HIN5 A0A1B6KT93 A0A1B6JRI3 A0A0K8T7K7 A0A3Q0IRY1 A0A1B6M0E3
Pubmed
EMBL
BABH01016071
BABH01016072
BABH01016073
BABH01016074
BABH01016075
ODYU01000597
+ More
SOQ35666.1 RSAL01000006 RVE54338.1 NWSH01000295 PCG77665.1 GDQN01008697 JAT82357.1 PCG77664.1 KQ459252 KPJ02147.1 PCG77667.1 AGBW02011469 OWR46596.1 KQ971319 EFA00314.2 GEZM01048768 JAV76421.1 NEVH01016330 PNF25599.1 PNF25600.1 KK852804 KDR16207.1 PNF25598.1 GL446605 EFN87552.1 GL439322 EFN67435.1 KQ977595 KYN01623.1 KQ979492 KYN21219.1 ADTU01006781 KQ414612 KOC69153.1 KQ982335 KYQ57448.1 GL887707 EGI70115.1 KQ436240 KOX67345.1 PYGN01001832 PSN32610.1 GEDC01000570 JAS36728.1 GEDC01020924 JAS16374.1 ACPB03004962 GECZ01016390 JAS53379.1 GFTR01008157 JAW08269.1 AMQN01013424 KB310211 ELT92237.1 GBGD01000449 JAC88440.1 GECL01002805 JAP03319.1 GG666590 EEN51736.1 GECU01033197 JAS74509.1 GEBQ01025304 JAT14673.1 GECU01006193 JAT01514.1 GBRD01004317 JAG61504.1 GEBQ01010612 JAT29365.1
SOQ35666.1 RSAL01000006 RVE54338.1 NWSH01000295 PCG77665.1 GDQN01008697 JAT82357.1 PCG77664.1 KQ459252 KPJ02147.1 PCG77667.1 AGBW02011469 OWR46596.1 KQ971319 EFA00314.2 GEZM01048768 JAV76421.1 NEVH01016330 PNF25599.1 PNF25600.1 KK852804 KDR16207.1 PNF25598.1 GL446605 EFN87552.1 GL439322 EFN67435.1 KQ977595 KYN01623.1 KQ979492 KYN21219.1 ADTU01006781 KQ414612 KOC69153.1 KQ982335 KYQ57448.1 GL887707 EGI70115.1 KQ436240 KOX67345.1 PYGN01001832 PSN32610.1 GEDC01000570 JAS36728.1 GEDC01020924 JAS16374.1 ACPB03004962 GECZ01016390 JAS53379.1 GFTR01008157 JAW08269.1 AMQN01013424 KB310211 ELT92237.1 GBGD01000449 JAC88440.1 GECL01002805 JAP03319.1 GG666590 EEN51736.1 GECU01033197 JAS74509.1 GEBQ01025304 JAT14673.1 GECU01006193 JAT01514.1 GBRD01004317 JAG61504.1 GEBQ01010612 JAT29365.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JRB8
A0A2H1V4G1
A0A3S2PKW8
A0A2A4K067
A0A1E1W5V1
A0A2A4K195
+ More
A0A194Q9F6 A0A2A4K0A2 A0A212EYM0 D6WEY6 A0A1Y1LTS8 A0A2J7QAL2 A0A2J7QAI0 A0A067R2I0 A0A2J7QAI1 E2B9W5 E2AGL5 A0A195CLP3 A0A151J8S2 A0A088AJL1 A0A158P1S7 A0A0L7RE78 A0A151XAX9 F4W6N9 A0A1W4WR76 A0A0M8ZMN9 A0A2P8XKS6 A0A1B6EFM1 A0A1B6CSG8 T1HCS5 A0A1B6FTB1 A0A224XJT2 R7TFG3 A0A069DX22 A0A0V0G5S2 C3Z7E0 A0A1B6HIN5 A0A1B6KT93 A0A1B6JRI3 A0A0K8T7K7 A0A3Q0IRY1 A0A1B6M0E3
A0A194Q9F6 A0A2A4K0A2 A0A212EYM0 D6WEY6 A0A1Y1LTS8 A0A2J7QAL2 A0A2J7QAI0 A0A067R2I0 A0A2J7QAI1 E2B9W5 E2AGL5 A0A195CLP3 A0A151J8S2 A0A088AJL1 A0A158P1S7 A0A0L7RE78 A0A151XAX9 F4W6N9 A0A1W4WR76 A0A0M8ZMN9 A0A2P8XKS6 A0A1B6EFM1 A0A1B6CSG8 T1HCS5 A0A1B6FTB1 A0A224XJT2 R7TFG3 A0A069DX22 A0A0V0G5S2 C3Z7E0 A0A1B6HIN5 A0A1B6KT93 A0A1B6JRI3 A0A0K8T7K7 A0A3Q0IRY1 A0A1B6M0E3
PDB
4BGJ
E-value=0.000153506,
Score=109
Ontologies
GO
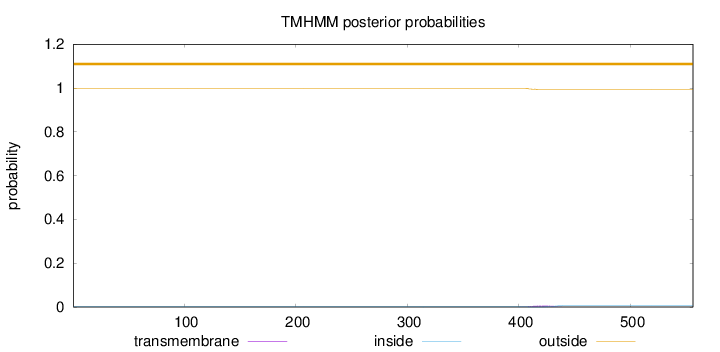
Topology
Length:
556
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14512
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00078
outside
1 - 556
Population Genetic Test Statistics
Pi
14.960505
Theta
16.11322
Tajima's D
-1.83591
CLR
0.63371
CSRT
0.0223488825558722
Interpretation
Uncertain
