Gene
KWMTBOMO06874
Pre Gene Modal
BGIBMGA012074
Annotation
PREDICTED:_retinol_dehydrogenase_11-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.826
Sequence
CDS
ATGTCTGTTATTATAGCATCTGCTGTTTTGAGCGCTGCCGCTTTTTATTGGTATGTAAACCGTGATAAACCAATCAAGCATATACTGAAAGAGATTCGGTACGCGTTGGACATGCAGGGCGCTGGGGCGGGTGGCCTTATTGACGACTGGATTAACGTGTCGAGGAACAGAATTGCGTTACCAGATGCGGCCGGAAAAGTTGCGGTCGTTACCGGCGGAGCGAGGGGCATCGGCACAGAAGTCATCAAGGGCTTACTCAAAGCAAACATGCGGGTGATAATCGGAATACGAAACCCGGAATTGGTGCATAAATTATCTGAATCAATGGAGAATGGTCAAAACTTGGAAGCTTACAAACTGGATTTGAAATCGTTAAAATCCGTCAAGGAATTTGCTGATAATGTACTGCATAAGTACCCAAAGATAAATTTACTAGTTAACAATGCTGGGATAATGTATGGGAAGTATGAATTGACTACAGACGGTTTTGAATCACAACTCGCTGTGAATCACCTGGGGCATTTTTATTTGACACACCTACTTTTGCCGGCTTTGAAGAAAGCTGGAAACTTATGTGAACCATCAAGGTTCATTAATGTGACGTCATGTGGCCACTATCCGGGACAAATATATTTTGATGATATTAACATGAAAGAGCATTATGATGAAACTGCAGCTTATGCACAATCCAAATTAGCTCAGTTAATGATGGCAAGATACATTAATAAGATTATGGAAGAGAATAATATTCCTGTCAAAGGATACTCAGTGCATCCCGGGATTGTCAACACTGATTTATTCGATGGTACAGTCTTCAAAAGAGTTTTTCCATGGGCTATGAAATTATTCTTCAAGACTCCAGAAAAGGGAGCCGTTTCTATACTCTTTGCGTGTTTCAATGAAGAATTGCAGAAGCGGGGAGGATTATACATTAGTAATTGTATTGAAGGGATCAGTAATAGATTTTCAAAGAATTTGGAATATCAAAAAATACTTTTTGAGCTATCCTGTCAATTAGTTGGAATTGATGCCGAGAACTTTGGAAATACTGAATAA
Protein
MSVIIASAVLSAAAFYWYVNRDKPIKHILKEIRYALDMQGAGAGGLIDDWINVSRNRIALPDAAGKVAVVTGGARGIGTEVIKGLLKANMRVIIGIRNPELVHKLSESMENGQNLEAYKLDLKSLKSVKEFADNVLHKYPKINLLVNNAGIMYGKYELTTDGFESQLAVNHLGHFYLTHLLLPALKKAGNLCEPSRFINVTSCGHYPGQIYFDDINMKEHYDETAAYAQSKLAQLMMARYINKIMEENNIPVKGYSVHPGIVNTDLFDGTVFKRVFPWAMKLFFKTPEKGAVSILFACFNEELQKRGGLYISNCIEGISNRFSKNLEYQKILFELSCQLVGIDAENFGNTE
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JRB4
A0A2A4K1X6
A0A0L7KMP1
A0A2H1V5W5
A0A3S2LUZ1
A0A2Z5U787
+ More
A0A1Q3G5H5 A0A194QR71 A0A194Q9R5 I4DJM0 A0A212ELP5 A0A023F1Q5 D2A0S5 A0A224XSD2 A0A067RDD8 A0A2J7R3B3 U4UJE2 T1I7Z2 J3JX50 A0A1Y1KWM7 W8BP01 A0A1A9WIU1 A0A2P8XV92 N6SRY5 A0A1A9V5D6 A0A1B0G7M5 A0A1Y1KWJ4 A0A1A9ZGK2 A0A1A9YKU8 A0A1B6DJW5 A0A182JCK3 Q16M73 A0A2M3Z1M9 A0A1I8Q4B6 A0A1B6GE33 A0A182L4U3 A0A2M4ATT2 A0A2M4ATK4 A0A2M4DMB0 A0A0Q9W3R2 A0A2M4DM74 B4LMT3 B4KN95 A0A1B0B7C5 A0A0Q9XBN3 A0A1B6I6L8 A0A0L0CN28 A0A2H8TZU1 E0W026 A0A1I8M3F6 A0A182TET0 A0NGL8 A0A182N3G4 A0A1L8E4R8 T1PIM4 A0A182QG16 A0A182FWA2 A0A1Q3G4Y5 A0A182US47 A0A1Q3G4V2 A0A034W7J8 A0A084WJ69 A0A1Q3G4X4 A0A1Q3G4T9 A0A0P5URS8 A0A182KBW6 A0A0P5M6Y6 A0A0P5FB50 A0A182W1U9 A0A2M4BUN4 A0A154NXX4 A0A1J1HL96 A0A182S6L4 A0A0P5S1K9 A0A182YL77 E2BHI2 A0A0P6F5S3 A0A0N8DXQ1 A0A0P6B9B8 A0A0A1WKF5 A0A0P6ELA1 A0A0P5KCF4 A0A0P5XWX9 A0A182P525 A0A0N8ABZ9 A0A0P5VKI7 A0A182MKR6 A0A0P6HUT1 B3MHL3 A0A182R7G5 A0A0P5RPH1 A0A0N8A5V5 A0A0P5RBI8 W5JWI1 A0A0K8U868 E9G0Q5 A0A232EN88 A0A1B0CDG0 C4WV09 B4MNQ4
A0A1Q3G5H5 A0A194QR71 A0A194Q9R5 I4DJM0 A0A212ELP5 A0A023F1Q5 D2A0S5 A0A224XSD2 A0A067RDD8 A0A2J7R3B3 U4UJE2 T1I7Z2 J3JX50 A0A1Y1KWM7 W8BP01 A0A1A9WIU1 A0A2P8XV92 N6SRY5 A0A1A9V5D6 A0A1B0G7M5 A0A1Y1KWJ4 A0A1A9ZGK2 A0A1A9YKU8 A0A1B6DJW5 A0A182JCK3 Q16M73 A0A2M3Z1M9 A0A1I8Q4B6 A0A1B6GE33 A0A182L4U3 A0A2M4ATT2 A0A2M4ATK4 A0A2M4DMB0 A0A0Q9W3R2 A0A2M4DM74 B4LMT3 B4KN95 A0A1B0B7C5 A0A0Q9XBN3 A0A1B6I6L8 A0A0L0CN28 A0A2H8TZU1 E0W026 A0A1I8M3F6 A0A182TET0 A0NGL8 A0A182N3G4 A0A1L8E4R8 T1PIM4 A0A182QG16 A0A182FWA2 A0A1Q3G4Y5 A0A182US47 A0A1Q3G4V2 A0A034W7J8 A0A084WJ69 A0A1Q3G4X4 A0A1Q3G4T9 A0A0P5URS8 A0A182KBW6 A0A0P5M6Y6 A0A0P5FB50 A0A182W1U9 A0A2M4BUN4 A0A154NXX4 A0A1J1HL96 A0A182S6L4 A0A0P5S1K9 A0A182YL77 E2BHI2 A0A0P6F5S3 A0A0N8DXQ1 A0A0P6B9B8 A0A0A1WKF5 A0A0P6ELA1 A0A0P5KCF4 A0A0P5XWX9 A0A182P525 A0A0N8ABZ9 A0A0P5VKI7 A0A182MKR6 A0A0P6HUT1 B3MHL3 A0A182R7G5 A0A0P5RPH1 A0A0N8A5V5 A0A0P5RBI8 W5JWI1 A0A0K8U868 E9G0Q5 A0A232EN88 A0A1B0CDG0 C4WV09 B4MNQ4
Pubmed
EMBL
BABH01016076
NWSH01000295
PCG77670.1
JTDY01008453
KOB64562.1
ODYU01000597
+ More
SOQ35664.1 RSAL01000006 RVE54337.1 AP017502 BBB06796.1 GEYN01000186 JAV44803.1 KQ461185 KPJ07470.1 KQ459252 KPJ02149.1 AK401488 BAM18110.1 AGBW02013998 OWR42412.1 GBBI01003778 JAC14934.1 KQ971338 EFA02555.1 GFTR01005076 JAW11350.1 KK852773 KDR16815.1 NEVH01007822 PNF35328.1 KB632349 ERL93222.1 ACPB03011620 BT127818 AEE62780.1 GEZM01071484 GEZM01071481 GEZM01071480 GEZM01071479 GEZM01071477 GEZM01071476 GEZM01071471 GEZM01071467 JAV65772.1 GAMC01003685 GAMC01003684 JAC02871.1 PYGN01001295 PSN35928.1 APGK01059088 APGK01059089 KB741292 ENN70404.1 CCAG010016887 GEZM01071483 GEZM01071478 GEZM01071474 GEZM01071472 GEZM01071470 GEZM01071468 JAV65783.1 GEDC01025017 GEDC01023787 GEDC01017777 GEDC01012711 GEDC01011314 JAS12281.1 JAS13511.1 JAS19521.1 JAS24587.1 JAS25984.1 CH477874 EAT35430.1 GGFM01001665 MBW22416.1 GECZ01009099 JAS60670.1 GGFK01010801 MBW44122.1 GGFK01010800 MBW44121.1 GGFL01014487 MBW78665.1 CH940648 KRF79737.1 GGFL01014488 MBW78666.1 EDW61024.2 CH933808 EDW09948.1 KRG04999.1 KRG05000.1 JXJN01009530 JXJN01009531 KRG05001.1 GECU01025134 JAS82572.1 JRES01000171 KNC33691.1 GFXV01007625 MBW19430.1 DS235855 EEB18982.1 AAAB01008986 EAL38858.2 GFDF01000460 JAV13624.1 KA648509 AFP63138.1 AXCN02001109 GFDL01000216 JAV34829.1 GFDL01000214 JAV34831.1 GAKP01009224 JAC49728.1 ATLV01023975 KE525347 KFB50263.1 GFDL01000226 JAV34819.1 GFDL01000228 JAV34817.1 GDIP01109307 JAL94407.1 GDIQ01159830 JAK91895.1 GDIP01149701 JAJ73701.1 GGFJ01007636 MBW56777.1 KQ434778 KZC04431.1 CVRI01000009 CRK88707.1 GDIQ01094250 JAL57476.1 GL448301 EFN84835.1 GDIQ01060973 JAN33764.1 GDIQ01081660 JAN13077.1 GDIP01018424 JAM85291.1 GBXI01015434 JAC98857.1 GDIQ01060974 JAN33763.1 GDIQ01191567 GDIP01077802 JAK60158.1 JAM25913.1 GDIP01066214 JAM37501.1 GDIP01162345 JAJ61057.1 GDIP01113563 JAL90151.1 AXCM01005693 GDIQ01038530 JAN56207.1 CH902619 EDV35849.1 KPU75859.1 GDIQ01097860 JAL53866.1 GDIP01179497 JAJ43905.1 GDIQ01108245 JAL43481.1 ADMH02000153 ETN67590.1 GDHF01029568 JAI22746.1 GL732528 EFX86945.1 NNAY01003205 OXU19796.1 AJWK01007802 AK341306 BAH71729.1 CH963848 EDW73743.1
SOQ35664.1 RSAL01000006 RVE54337.1 AP017502 BBB06796.1 GEYN01000186 JAV44803.1 KQ461185 KPJ07470.1 KQ459252 KPJ02149.1 AK401488 BAM18110.1 AGBW02013998 OWR42412.1 GBBI01003778 JAC14934.1 KQ971338 EFA02555.1 GFTR01005076 JAW11350.1 KK852773 KDR16815.1 NEVH01007822 PNF35328.1 KB632349 ERL93222.1 ACPB03011620 BT127818 AEE62780.1 GEZM01071484 GEZM01071481 GEZM01071480 GEZM01071479 GEZM01071477 GEZM01071476 GEZM01071471 GEZM01071467 JAV65772.1 GAMC01003685 GAMC01003684 JAC02871.1 PYGN01001295 PSN35928.1 APGK01059088 APGK01059089 KB741292 ENN70404.1 CCAG010016887 GEZM01071483 GEZM01071478 GEZM01071474 GEZM01071472 GEZM01071470 GEZM01071468 JAV65783.1 GEDC01025017 GEDC01023787 GEDC01017777 GEDC01012711 GEDC01011314 JAS12281.1 JAS13511.1 JAS19521.1 JAS24587.1 JAS25984.1 CH477874 EAT35430.1 GGFM01001665 MBW22416.1 GECZ01009099 JAS60670.1 GGFK01010801 MBW44122.1 GGFK01010800 MBW44121.1 GGFL01014487 MBW78665.1 CH940648 KRF79737.1 GGFL01014488 MBW78666.1 EDW61024.2 CH933808 EDW09948.1 KRG04999.1 KRG05000.1 JXJN01009530 JXJN01009531 KRG05001.1 GECU01025134 JAS82572.1 JRES01000171 KNC33691.1 GFXV01007625 MBW19430.1 DS235855 EEB18982.1 AAAB01008986 EAL38858.2 GFDF01000460 JAV13624.1 KA648509 AFP63138.1 AXCN02001109 GFDL01000216 JAV34829.1 GFDL01000214 JAV34831.1 GAKP01009224 JAC49728.1 ATLV01023975 KE525347 KFB50263.1 GFDL01000226 JAV34819.1 GFDL01000228 JAV34817.1 GDIP01109307 JAL94407.1 GDIQ01159830 JAK91895.1 GDIP01149701 JAJ73701.1 GGFJ01007636 MBW56777.1 KQ434778 KZC04431.1 CVRI01000009 CRK88707.1 GDIQ01094250 JAL57476.1 GL448301 EFN84835.1 GDIQ01060973 JAN33764.1 GDIQ01081660 JAN13077.1 GDIP01018424 JAM85291.1 GBXI01015434 JAC98857.1 GDIQ01060974 JAN33763.1 GDIQ01191567 GDIP01077802 JAK60158.1 JAM25913.1 GDIP01066214 JAM37501.1 GDIP01162345 JAJ61057.1 GDIP01113563 JAL90151.1 AXCM01005693 GDIQ01038530 JAN56207.1 CH902619 EDV35849.1 KPU75859.1 GDIQ01097860 JAL53866.1 GDIP01179497 JAJ43905.1 GDIQ01108245 JAL43481.1 ADMH02000153 ETN67590.1 GDHF01029568 JAI22746.1 GL732528 EFX86945.1 NNAY01003205 OXU19796.1 AJWK01007802 AK341306 BAH71729.1 CH963848 EDW73743.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000053240
UP000053268
+ More
UP000007151 UP000007266 UP000027135 UP000235965 UP000030742 UP000015103 UP000091820 UP000245037 UP000019118 UP000078200 UP000092444 UP000092445 UP000092443 UP000075880 UP000008820 UP000095300 UP000075882 UP000008792 UP000009192 UP000092460 UP000037069 UP000009046 UP000095301 UP000075902 UP000007062 UP000075884 UP000075886 UP000069272 UP000075903 UP000030765 UP000075881 UP000075920 UP000076502 UP000183832 UP000075901 UP000076408 UP000008237 UP000075885 UP000075883 UP000007801 UP000075900 UP000000673 UP000000305 UP000215335 UP000092461 UP000007798
UP000007151 UP000007266 UP000027135 UP000235965 UP000030742 UP000015103 UP000091820 UP000245037 UP000019118 UP000078200 UP000092444 UP000092445 UP000092443 UP000075880 UP000008820 UP000095300 UP000075882 UP000008792 UP000009192 UP000092460 UP000037069 UP000009046 UP000095301 UP000075902 UP000007062 UP000075884 UP000075886 UP000069272 UP000075903 UP000030765 UP000075881 UP000075920 UP000076502 UP000183832 UP000075901 UP000076408 UP000008237 UP000075885 UP000075883 UP000007801 UP000075900 UP000000673 UP000000305 UP000215335 UP000092461 UP000007798
PRIDE
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JRB4
A0A2A4K1X6
A0A0L7KMP1
A0A2H1V5W5
A0A3S2LUZ1
A0A2Z5U787
+ More
A0A1Q3G5H5 A0A194QR71 A0A194Q9R5 I4DJM0 A0A212ELP5 A0A023F1Q5 D2A0S5 A0A224XSD2 A0A067RDD8 A0A2J7R3B3 U4UJE2 T1I7Z2 J3JX50 A0A1Y1KWM7 W8BP01 A0A1A9WIU1 A0A2P8XV92 N6SRY5 A0A1A9V5D6 A0A1B0G7M5 A0A1Y1KWJ4 A0A1A9ZGK2 A0A1A9YKU8 A0A1B6DJW5 A0A182JCK3 Q16M73 A0A2M3Z1M9 A0A1I8Q4B6 A0A1B6GE33 A0A182L4U3 A0A2M4ATT2 A0A2M4ATK4 A0A2M4DMB0 A0A0Q9W3R2 A0A2M4DM74 B4LMT3 B4KN95 A0A1B0B7C5 A0A0Q9XBN3 A0A1B6I6L8 A0A0L0CN28 A0A2H8TZU1 E0W026 A0A1I8M3F6 A0A182TET0 A0NGL8 A0A182N3G4 A0A1L8E4R8 T1PIM4 A0A182QG16 A0A182FWA2 A0A1Q3G4Y5 A0A182US47 A0A1Q3G4V2 A0A034W7J8 A0A084WJ69 A0A1Q3G4X4 A0A1Q3G4T9 A0A0P5URS8 A0A182KBW6 A0A0P5M6Y6 A0A0P5FB50 A0A182W1U9 A0A2M4BUN4 A0A154NXX4 A0A1J1HL96 A0A182S6L4 A0A0P5S1K9 A0A182YL77 E2BHI2 A0A0P6F5S3 A0A0N8DXQ1 A0A0P6B9B8 A0A0A1WKF5 A0A0P6ELA1 A0A0P5KCF4 A0A0P5XWX9 A0A182P525 A0A0N8ABZ9 A0A0P5VKI7 A0A182MKR6 A0A0P6HUT1 B3MHL3 A0A182R7G5 A0A0P5RPH1 A0A0N8A5V5 A0A0P5RBI8 W5JWI1 A0A0K8U868 E9G0Q5 A0A232EN88 A0A1B0CDG0 C4WV09 B4MNQ4
A0A1Q3G5H5 A0A194QR71 A0A194Q9R5 I4DJM0 A0A212ELP5 A0A023F1Q5 D2A0S5 A0A224XSD2 A0A067RDD8 A0A2J7R3B3 U4UJE2 T1I7Z2 J3JX50 A0A1Y1KWM7 W8BP01 A0A1A9WIU1 A0A2P8XV92 N6SRY5 A0A1A9V5D6 A0A1B0G7M5 A0A1Y1KWJ4 A0A1A9ZGK2 A0A1A9YKU8 A0A1B6DJW5 A0A182JCK3 Q16M73 A0A2M3Z1M9 A0A1I8Q4B6 A0A1B6GE33 A0A182L4U3 A0A2M4ATT2 A0A2M4ATK4 A0A2M4DMB0 A0A0Q9W3R2 A0A2M4DM74 B4LMT3 B4KN95 A0A1B0B7C5 A0A0Q9XBN3 A0A1B6I6L8 A0A0L0CN28 A0A2H8TZU1 E0W026 A0A1I8M3F6 A0A182TET0 A0NGL8 A0A182N3G4 A0A1L8E4R8 T1PIM4 A0A182QG16 A0A182FWA2 A0A1Q3G4Y5 A0A182US47 A0A1Q3G4V2 A0A034W7J8 A0A084WJ69 A0A1Q3G4X4 A0A1Q3G4T9 A0A0P5URS8 A0A182KBW6 A0A0P5M6Y6 A0A0P5FB50 A0A182W1U9 A0A2M4BUN4 A0A154NXX4 A0A1J1HL96 A0A182S6L4 A0A0P5S1K9 A0A182YL77 E2BHI2 A0A0P6F5S3 A0A0N8DXQ1 A0A0P6B9B8 A0A0A1WKF5 A0A0P6ELA1 A0A0P5KCF4 A0A0P5XWX9 A0A182P525 A0A0N8ABZ9 A0A0P5VKI7 A0A182MKR6 A0A0P6HUT1 B3MHL3 A0A182R7G5 A0A0P5RPH1 A0A0N8A5V5 A0A0P5RBI8 W5JWI1 A0A0K8U868 E9G0Q5 A0A232EN88 A0A1B0CDG0 C4WV09 B4MNQ4
PDB
3RD5
E-value=2.51396e-28,
Score=312
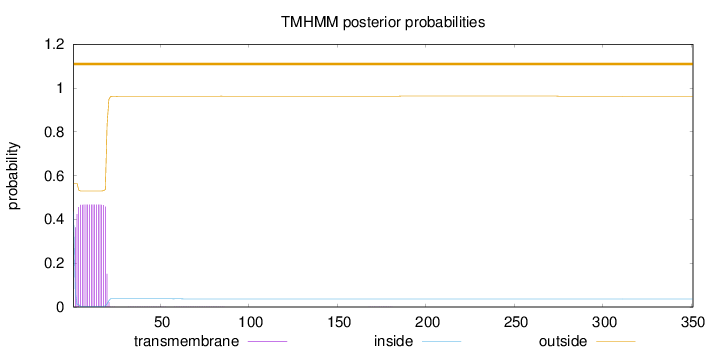
Ontologies
Topology
Length:
351
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.44451999999999
Exp number, first 60 AAs:
8.42504
Total prob of N-in:
0.43555
outside
1 - 351
Population Genetic Test Statistics
Pi
17.621168
Theta
14.794717
Tajima's D
0.340337
CLR
0.245809
CSRT
0.474976251187441
Interpretation
Uncertain
