Gene
KWMTBOMO06872 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012082
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.989
Sequence
CDS
ATGAGATCGTGCTTATTTCGAAAAAGAGGTAGTGATGACGAAAGGGAACCTTTGCTTCCCGCGATCCCTGTATCCGAATCAAGTTCAAATACCGGAAATGAGTGTGCTAGCGAGACAAACACAAACGGCAGGATGGCCAATCTCGGTGTCTCTCAAACAGAACTGGTTGGTCCTCGGGGAGATTCTGGTCGCAAGTTACCTCAATACATAGCTGCTTTAGCAGCTACCCTTGGAGCACTAGCTGCTGGAACAATGCTTGGTTGGTCTTCGCCAGTTGTGTTCAAAATAACACAACCAAACAATACAGACTACAATTTTGACATCAGTGAAACTGAAGGAAGCTGGATTGGGTCTGTCATCAACTTGGGTGCTGCTGCAATTTGTTTTCCAATTGGCCTGGTCATGGATGCTATTGGACGAAAAAAAACTATGCTGTTTCTCATCCTCCCGTTCACTTTGGGCTGGCTTCTTATAACTTTTGGTACGAGTGTCGGTATGCTCATAGGCGGAAGACTCATTACCGGTATTGCAGGAGGAGCTTTCTGTGTCACCGCACCGGCATACACTAGTGAAATAGCCCAAGACTCCATCAGAGGTACCCTAGGGAGCTACTTCCAACTGATGATCACAGTTGGTATTCTATTTGCTTATGCAGTGGGTAGTTACACATCAGTGTTCCTTTTCAATATTCTTTGCACTCTGATACCTATTGTCTTTGGTATTGTGTTCTTTTTCATGCCTGAAAGTCCAAAATTTTTGGTTGTAAAAAACAGAAACGATGAAGCTCGAGAAGCACTAATTAAACTACGCGGTACCAATTACGATGTTGACTACGAGCTGGACTCTTTGAAACTGAGCGCTGAAGAAGCACAGAACAATCCAGTTTCATTTGTTTCGGCTATTACGAAGAAAACGTCTATTAAAGCGATAATAATTTGTTACGCACTGATGATATTCCAACAGCTCTCTGGTATAAATGCGGTCATTTTTAACACCTCATCGATTTTCGCTGCTGCCGGTGCCACAATTCCGGCCGCGATCGCGACCATTATCATCGGCGTCATCCAAGTAATCGCCACATTTGTATCCAGTTTGGTTGTCGACAAATTAGGCCGACGTATTTTGCTTCTCTTCTCCGCTTTGGTTATGTGCTTGTGTTCGACGGCTTTGGGGGTGTTTTTCTTCTTACAAAGTACGCACGGAGAAAATTCTGATATAGTTCAAAGTCTTTTCTGGTTACCTCTTTTGTCTTTGTCTCTTTTCATCATCGCGTTCTCTCTTGGTTTCGGTCCGATCCCGTGGATGATGGCTGGCGATCTCTGCAATATTGACATAAAGGCGTTCGTCGGTTCCACTGCCGGGACATTGAACTGGCTGCTTAGTTTTACGGTGACCAGCACCTTCCTTGCCTTGAATACTGCTATTGGATCTGGTCAGGTATTCTGGATGTTCGCAGGCATCATGTTGATTGGTTTTGTATTCATTTTCTTCGTAATACCGGAGACAAAAGGCAAGAGCCTTCAAGAAATTCAAGTGATGTTAGGAGCGACACCACAAGATAGAAACGTAGAAGACAAAAAGTGA
Protein
MRSCLFRKRGSDDEREPLLPAIPVSESSSNTGNECASETNTNGRMANLGVSQTELVGPRGDSGRKLPQYIAALAATLGALAAGTMLGWSSPVVFKITQPNNTDYNFDISETEGSWIGSVINLGAAAICFPIGLVMDAIGRKKTMLFLILPFTLGWLLITFGTSVGMLIGGRLITGIAGGAFCVTAPAYTSEIAQDSIRGTLGSYFQLMITVGILFAYAVGSYTSVFLFNILCTLIPIVFGIVFFFMPESPKFLVVKNRNDEAREALIKLRGTNYDVDYELDSLKLSAEEAQNNPVSFVSAITKKTSIKAIIICYALMIFQQLSGINAVIFNTSSIFAAAGATIPAAIATIIIGVIQVIATFVSSLVVDKLGRRILLLFSALVMCLCSTALGVFFFLQSTHGENSDIVQSLFWLPLLSLSLFIIAFSLGFGPIPWMMAGDLCNIDIKAFVGSTAGTLNWLLSFTVTSTFLALNTAIGSGQVFWMFAGIMLIGFVFIFFVIPETKGKSLQEIQVMLGATPQDRNVEDKK
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9JRC2
A0A2H1WM07
A0A194Q9H4
A0A194QPN7
S4PA10
A0A2A4K8N6
+ More
A0A2A4K8F1 A0A0L7KYV1 A0A212EQ73 A0A2Z5U777 A0A3B0JV95 A0A0R3P5D7 U5EQL0 A0A3B0J6A5 B4ML33 A0A0Q9XD79 B3M9B8 B5DQ85 A0A3B0J5Y8 A0A1W4UHA6 B4HG70 A0A0Q9WVX8 A0A0J9RVP2 B4L0T8 A0A0R3P588 A0A0P8YC58 A0A0Q9XE05 Q9VU17 B3NCV6 A0A1W4UVY7 A0A182W4W6 A0A182KB78 A0A182JJ49 A0A0J9UK27 B4IZM8 A0A1J1IWB4 A0A182XLV5 A0A336LQF3 A0A182UR48 B4LBJ1 Q7PSV5 A0A0A1WTN4 T1PCW2 A0A182MEP6 A0A1I8M2U2 A0A182RP90 A0A182LIB9 Q8IQH6 A0A0Q9WK01 B4PGV6 A0A0M3QWT3 A0A0K8UNM6 A0A0Q5U452 A0A182IDU0 A0A0R1E2J5 A0A182SZA1 A0A2M4BL39 A0A182UJT1 A0A182Y502 D3TR19 A0A1I8P3B0 A0A0A1WG49 A0A1I8P360 A0A182QX63 B0X8T3 A0A034VPA7 F5HJ38 F5HJ37 A0A1Q3FJI4 A0A1L8DF43 Q17EH6 Q16ZD3 A0A2M3Z9X6 A0A182PI25 A0A1B0F9G0 A0A232ERM9 A0A182N407 A0A084VLL6 A0A1Y1NE39 A0A1Y1N7N8 A0A1A9ZKQ0 W8B5S9 A0A023ET97 A0A023EU90 A0A182GCG2 A0A1S4F5X0 A0A067RCA0 A0A1Y0AWQ1 A0A0J7N829 B4QRS6 W0FQU0 A0A139WD56 T1GCU9 A0A1L8DIL6 V5GV83 A0A0T6B7J6 Q17EH5 A0A151XG97 A0A182GCF4 A0A1A9UFU3
A0A2A4K8F1 A0A0L7KYV1 A0A212EQ73 A0A2Z5U777 A0A3B0JV95 A0A0R3P5D7 U5EQL0 A0A3B0J6A5 B4ML33 A0A0Q9XD79 B3M9B8 B5DQ85 A0A3B0J5Y8 A0A1W4UHA6 B4HG70 A0A0Q9WVX8 A0A0J9RVP2 B4L0T8 A0A0R3P588 A0A0P8YC58 A0A0Q9XE05 Q9VU17 B3NCV6 A0A1W4UVY7 A0A182W4W6 A0A182KB78 A0A182JJ49 A0A0J9UK27 B4IZM8 A0A1J1IWB4 A0A182XLV5 A0A336LQF3 A0A182UR48 B4LBJ1 Q7PSV5 A0A0A1WTN4 T1PCW2 A0A182MEP6 A0A1I8M2U2 A0A182RP90 A0A182LIB9 Q8IQH6 A0A0Q9WK01 B4PGV6 A0A0M3QWT3 A0A0K8UNM6 A0A0Q5U452 A0A182IDU0 A0A0R1E2J5 A0A182SZA1 A0A2M4BL39 A0A182UJT1 A0A182Y502 D3TR19 A0A1I8P3B0 A0A0A1WG49 A0A1I8P360 A0A182QX63 B0X8T3 A0A034VPA7 F5HJ38 F5HJ37 A0A1Q3FJI4 A0A1L8DF43 Q17EH6 Q16ZD3 A0A2M3Z9X6 A0A182PI25 A0A1B0F9G0 A0A232ERM9 A0A182N407 A0A084VLL6 A0A1Y1NE39 A0A1Y1N7N8 A0A1A9ZKQ0 W8B5S9 A0A023ET97 A0A023EU90 A0A182GCG2 A0A1S4F5X0 A0A067RCA0 A0A1Y0AWQ1 A0A0J7N829 B4QRS6 W0FQU0 A0A139WD56 T1GCU9 A0A1L8DIL6 V5GV83 A0A0T6B7J6 Q17EH5 A0A151XG97 A0A182GCF4 A0A1A9UFU3
Pubmed
19121390
26354079
23622113
26227816
22118469
15632085
+ More
17994087 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 25830018 25315136 20966253 17550304 25244985 20353571 25348373 17510324 28648823 24438588 28004739 24495485 24945155 26483478 24845553 28341416 24391959 18362917 19820115
17994087 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 25830018 25315136 20966253 17550304 25244985 20353571 25348373 17510324 28648823 24438588 28004739 24495485 24945155 26483478 24845553 28341416 24391959 18362917 19820115
EMBL
BABH01016085
BABH01016086
ODYU01009579
SOQ54105.1
KQ459252
KPJ02153.1
+ More
KQ461185 KPJ07467.1 GAIX01003464 JAA89096.1 NWSH01000052 PCG80173.1 PCG80174.1 JTDY01004240 KOB68407.1 AGBW02013309 OWR43617.1 AP017502 BBB06794.1 OUUW01000002 SPP77296.1 CH379069 KRT08322.1 GANO01004191 JAB55680.1 SPP77295.1 CH963847 EDW73091.2 CH933809 KRG06534.1 CH902618 EDV41131.1 EDY73816.1 SPP77297.1 CH480815 EDW41316.1 CH940647 KRF85118.1 CM002912 KMY99309.1 EDW19188.1 KRT08321.1 KPU78958.1 KPU78959.1 KPU78960.1 KRG06533.1 AE014296 AY119564 AAF49874.1 AAM50218.1 CH954178 EDV51612.1 KMY99310.1 CH916366 EDV97803.1 CVRI01000063 CRL04571.1 UFQS01000107 UFQT01000107 SSW99705.1 SSX20085.1 EDW70801.1 AAAB01008811 EAA04970.6 GBXI01012080 JAD02212.1 KA646494 AFP61123.1 AXCM01004396 AAN11863.1 AAN11864.1 KRF85117.1 CM000159 EDW94345.1 CP012525 ALC44659.1 GDHF01024196 GDHF01015258 GDHF01004851 JAI28118.1 JAI37056.1 JAI47463.1 KQS43871.1 APCN01004802 KRK01780.1 GGFJ01004530 MBW53671.1 EZ423871 ADD20147.1 GBXI01016924 JAC97367.1 AXCN02002612 DS232501 EDS42675.1 GAKP01015559 GAKP01015558 GAKP01015557 JAC43393.1 EGK96298.1 EGK96299.1 GFDL01007265 JAV27780.1 GFDF01009023 JAV05061.1 CH477282 EAT44903.1 CH477492 EAT40014.1 GGFM01004529 MBW25280.1 CCAG010012835 NNAY01002587 OXU20982.1 ATLV01014522 KE524974 KFB38860.1 GEZM01010698 GEZM01010697 JAV93897.1 GEZM01010696 JAV93901.1 GAMC01021381 JAB85174.1 GAPW01001080 JAC12518.1 GAPW01001122 JAC12476.1 JXUM01054970 JXUM01054971 KQ561845 KXJ77365.1 KK852731 KDR17493.1 KY921823 ART29412.1 LBMM01008561 KMQ88790.1 CM000363 EDX10290.1 KF803261 KF803262 AHF27415.1 AHF27416.1 KQ971361 KYB25889.1 CAQQ02064192 GFDF01007788 JAV06296.1 GALX01004323 JAB64143.1 LJIG01009574 KRT82807.1 EAT44904.1 KQ982174 KYQ59377.1 JXUM01054954 KXJ77357.1
KQ461185 KPJ07467.1 GAIX01003464 JAA89096.1 NWSH01000052 PCG80173.1 PCG80174.1 JTDY01004240 KOB68407.1 AGBW02013309 OWR43617.1 AP017502 BBB06794.1 OUUW01000002 SPP77296.1 CH379069 KRT08322.1 GANO01004191 JAB55680.1 SPP77295.1 CH963847 EDW73091.2 CH933809 KRG06534.1 CH902618 EDV41131.1 EDY73816.1 SPP77297.1 CH480815 EDW41316.1 CH940647 KRF85118.1 CM002912 KMY99309.1 EDW19188.1 KRT08321.1 KPU78958.1 KPU78959.1 KPU78960.1 KRG06533.1 AE014296 AY119564 AAF49874.1 AAM50218.1 CH954178 EDV51612.1 KMY99310.1 CH916366 EDV97803.1 CVRI01000063 CRL04571.1 UFQS01000107 UFQT01000107 SSW99705.1 SSX20085.1 EDW70801.1 AAAB01008811 EAA04970.6 GBXI01012080 JAD02212.1 KA646494 AFP61123.1 AXCM01004396 AAN11863.1 AAN11864.1 KRF85117.1 CM000159 EDW94345.1 CP012525 ALC44659.1 GDHF01024196 GDHF01015258 GDHF01004851 JAI28118.1 JAI37056.1 JAI47463.1 KQS43871.1 APCN01004802 KRK01780.1 GGFJ01004530 MBW53671.1 EZ423871 ADD20147.1 GBXI01016924 JAC97367.1 AXCN02002612 DS232501 EDS42675.1 GAKP01015559 GAKP01015558 GAKP01015557 JAC43393.1 EGK96298.1 EGK96299.1 GFDL01007265 JAV27780.1 GFDF01009023 JAV05061.1 CH477282 EAT44903.1 CH477492 EAT40014.1 GGFM01004529 MBW25280.1 CCAG010012835 NNAY01002587 OXU20982.1 ATLV01014522 KE524974 KFB38860.1 GEZM01010698 GEZM01010697 JAV93897.1 GEZM01010696 JAV93901.1 GAMC01021381 JAB85174.1 GAPW01001080 JAC12518.1 GAPW01001122 JAC12476.1 JXUM01054970 JXUM01054971 KQ561845 KXJ77365.1 KK852731 KDR17493.1 KY921823 ART29412.1 LBMM01008561 KMQ88790.1 CM000363 EDX10290.1 KF803261 KF803262 AHF27415.1 AHF27416.1 KQ971361 KYB25889.1 CAQQ02064192 GFDF01007788 JAV06296.1 GALX01004323 JAB64143.1 LJIG01009574 KRT82807.1 EAT44904.1 KQ982174 KYQ59377.1 JXUM01054954 KXJ77357.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000037510
UP000007151
+ More
UP000268350 UP000001819 UP000007798 UP000009192 UP000007801 UP000192221 UP000001292 UP000008792 UP000000803 UP000008711 UP000075920 UP000075881 UP000075880 UP000001070 UP000183832 UP000076407 UP000075903 UP000007062 UP000075883 UP000095301 UP000075900 UP000075882 UP000002282 UP000092553 UP000075840 UP000075901 UP000075902 UP000076408 UP000095300 UP000075886 UP000002320 UP000008820 UP000075885 UP000092444 UP000215335 UP000075884 UP000030765 UP000092445 UP000069940 UP000249989 UP000027135 UP000036403 UP000000304 UP000007266 UP000015102 UP000075809 UP000078200
UP000268350 UP000001819 UP000007798 UP000009192 UP000007801 UP000192221 UP000001292 UP000008792 UP000000803 UP000008711 UP000075920 UP000075881 UP000075880 UP000001070 UP000183832 UP000076407 UP000075903 UP000007062 UP000075883 UP000095301 UP000075900 UP000075882 UP000002282 UP000092553 UP000075840 UP000075901 UP000075902 UP000076408 UP000095300 UP000075886 UP000002320 UP000008820 UP000075885 UP000092444 UP000215335 UP000075884 UP000030765 UP000092445 UP000069940 UP000249989 UP000027135 UP000036403 UP000000304 UP000007266 UP000015102 UP000075809 UP000078200
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JRC2
A0A2H1WM07
A0A194Q9H4
A0A194QPN7
S4PA10
A0A2A4K8N6
+ More
A0A2A4K8F1 A0A0L7KYV1 A0A212EQ73 A0A2Z5U777 A0A3B0JV95 A0A0R3P5D7 U5EQL0 A0A3B0J6A5 B4ML33 A0A0Q9XD79 B3M9B8 B5DQ85 A0A3B0J5Y8 A0A1W4UHA6 B4HG70 A0A0Q9WVX8 A0A0J9RVP2 B4L0T8 A0A0R3P588 A0A0P8YC58 A0A0Q9XE05 Q9VU17 B3NCV6 A0A1W4UVY7 A0A182W4W6 A0A182KB78 A0A182JJ49 A0A0J9UK27 B4IZM8 A0A1J1IWB4 A0A182XLV5 A0A336LQF3 A0A182UR48 B4LBJ1 Q7PSV5 A0A0A1WTN4 T1PCW2 A0A182MEP6 A0A1I8M2U2 A0A182RP90 A0A182LIB9 Q8IQH6 A0A0Q9WK01 B4PGV6 A0A0M3QWT3 A0A0K8UNM6 A0A0Q5U452 A0A182IDU0 A0A0R1E2J5 A0A182SZA1 A0A2M4BL39 A0A182UJT1 A0A182Y502 D3TR19 A0A1I8P3B0 A0A0A1WG49 A0A1I8P360 A0A182QX63 B0X8T3 A0A034VPA7 F5HJ38 F5HJ37 A0A1Q3FJI4 A0A1L8DF43 Q17EH6 Q16ZD3 A0A2M3Z9X6 A0A182PI25 A0A1B0F9G0 A0A232ERM9 A0A182N407 A0A084VLL6 A0A1Y1NE39 A0A1Y1N7N8 A0A1A9ZKQ0 W8B5S9 A0A023ET97 A0A023EU90 A0A182GCG2 A0A1S4F5X0 A0A067RCA0 A0A1Y0AWQ1 A0A0J7N829 B4QRS6 W0FQU0 A0A139WD56 T1GCU9 A0A1L8DIL6 V5GV83 A0A0T6B7J6 Q17EH5 A0A151XG97 A0A182GCF4 A0A1A9UFU3
A0A2A4K8F1 A0A0L7KYV1 A0A212EQ73 A0A2Z5U777 A0A3B0JV95 A0A0R3P5D7 U5EQL0 A0A3B0J6A5 B4ML33 A0A0Q9XD79 B3M9B8 B5DQ85 A0A3B0J5Y8 A0A1W4UHA6 B4HG70 A0A0Q9WVX8 A0A0J9RVP2 B4L0T8 A0A0R3P588 A0A0P8YC58 A0A0Q9XE05 Q9VU17 B3NCV6 A0A1W4UVY7 A0A182W4W6 A0A182KB78 A0A182JJ49 A0A0J9UK27 B4IZM8 A0A1J1IWB4 A0A182XLV5 A0A336LQF3 A0A182UR48 B4LBJ1 Q7PSV5 A0A0A1WTN4 T1PCW2 A0A182MEP6 A0A1I8M2U2 A0A182RP90 A0A182LIB9 Q8IQH6 A0A0Q9WK01 B4PGV6 A0A0M3QWT3 A0A0K8UNM6 A0A0Q5U452 A0A182IDU0 A0A0R1E2J5 A0A182SZA1 A0A2M4BL39 A0A182UJT1 A0A182Y502 D3TR19 A0A1I8P3B0 A0A0A1WG49 A0A1I8P360 A0A182QX63 B0X8T3 A0A034VPA7 F5HJ38 F5HJ37 A0A1Q3FJI4 A0A1L8DF43 Q17EH6 Q16ZD3 A0A2M3Z9X6 A0A182PI25 A0A1B0F9G0 A0A232ERM9 A0A182N407 A0A084VLL6 A0A1Y1NE39 A0A1Y1N7N8 A0A1A9ZKQ0 W8B5S9 A0A023ET97 A0A023EU90 A0A182GCG2 A0A1S4F5X0 A0A067RCA0 A0A1Y0AWQ1 A0A0J7N829 B4QRS6 W0FQU0 A0A139WD56 T1GCU9 A0A1L8DIL6 V5GV83 A0A0T6B7J6 Q17EH5 A0A151XG97 A0A182GCF4 A0A1A9UFU3
PDB
6H7D
E-value=3.1185e-31,
Score=339
Ontologies
GO
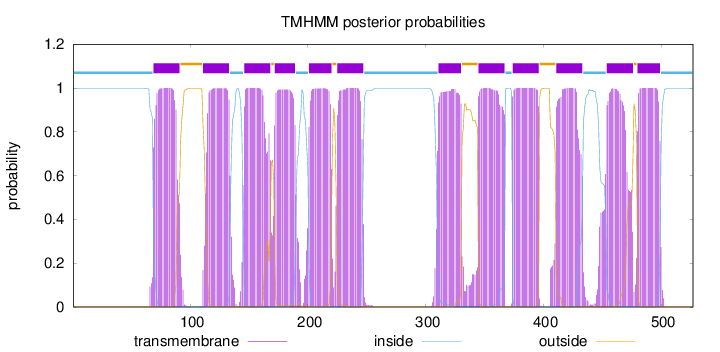
Topology
Length:
527
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
261.99779
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99736
inside
1 - 68
TMhelix
69 - 91
outside
92 - 110
TMhelix
111 - 133
inside
134 - 145
TMhelix
146 - 168
outside
169 - 171
TMhelix
172 - 189
inside
190 - 200
TMhelix
201 - 220
outside
221 - 224
TMhelix
225 - 247
inside
248 - 310
TMhelix
311 - 330
outside
331 - 344
TMhelix
345 - 367
inside
368 - 373
TMhelix
374 - 396
outside
397 - 410
TMhelix
411 - 433
inside
434 - 453
TMhelix
454 - 476
outside
477 - 479
TMhelix
480 - 499
inside
500 - 527
Population Genetic Test Statistics
Pi
13.857348
Theta
14.499245
Tajima's D
-0.765868
CLR
0.835135
CSRT
0.185390730463477
Interpretation
Uncertain
