Gene
KWMTBOMO06855
Pre Gene Modal
BGIBMGA012088
Annotation
cysteine_sulfinic_acid_decarboxylase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.516
Sequence
CDS
ATGCCTGCCGATTCAGATCTTATAGTCGCCGAGAGAGAGGACTTGGGCATTGATAGAACCCTCTACAAGTCACTCTCAGAGAGATCTAAGCATGAAGATTTTATCAGGCGGGCAGTGGATTTGCTGGTTGAAAGGGTAGTGTTCGGAAGGTCCCTGAGGACTTCTAAGGTGGTGGAATGGGCTGTACCGTCGGAGATCAAGAAAGCCATCGACTTGAAACCTAGAGATGGGCCCATTTCTCACGATGAACTTTTGGGACTCATGGCTGATGTGGCCCGATATTCCGTGAACACAGCCCATCCGTATTTCGTAAATCAGTTGTATTCATCCGTGGACCCGTATGGCTTGGTCGGACAATGGCTGACAGATGCCCTCAATCCAAGCGTGTATACCTATGAAGTTGCACCAGTTTTTACTCTAATGGAGGAAGAAGTGCTAAAGGAGATGAGGGTACTCGTTGGATGGAAAGATGGAGAAGGTGATGGTATATTTTGCCCGGGTGGTTCCATTGCCAACGGTTATGCCATCAGCTGCGCTAGATTCCATTTCTATCCTGAAATAAAGACAAAAGGCGTATACGCTGTACCGAAACTGACGCTATACACATCGGAGCTAGCACATTACTCGACGAAAAAATTGGCCGCATTCATGGGGATAGGAGATGAAAACTGCGTTTTAATAAAAACCGATAAATACGGTAAAATCGATGTTGAAGATTTAGAGGCTAAAATTGTGGAGGGCATCGAGGAGGGCGCTGCTCCATTCCTCGTCACAGCCACCGCTGGTACCACAGTCTTTGGTGCTTTCGACCCATTAGTCCCCATAGCGGCGCTCTGCAAGAAATACAATCTCTGGCTTCATGTTGACGCCGCTTGGGGTGGTGGAGCCTTAATGTCGAAGAAACATAGGCATCTGTTGAATGGAATTGAATTGGCGGATTCAGTCACGTGGAATCCCCACAAGCTACTGGCTGCTCCTCAACAATGCTCCACGTTCCTAATTAGACACAAGAACGTTCTCAAGGAAGGACACTCGTGCAATGCGAAGTACCTCTTCCAGAAGGACAAATTCTACGACACATCATACGATACGGGGGACAAACATATCCAGTGCGGTCGACGGGCGGATGTCCTCAAATTCTGGTTCATGTGGAAGGCCAAAGGCTCGGACGGCTTCGAGAACCACATCGATACGCTATTCGATAACGCGCGCTTCTTCTTGGAACAGATTCGTAATCGGGAAGGCTTCGAATTGGTCATTGAAAAACCGGAATGCACGAACATAATGTTCTGGTACGTACCGCGTTGCCTTCGCGGCTGCGAAAACGAGTCGGACTACAGAGAAAGGCTCCACAAGGTTGCCCCTAAAATTAAAGAATTAATGATAAAGGAAGGAAGTATGATGGTAACTTATCAGCCTCAAGGAGATCTTGTGAATTTCTTCCGTATCGTGTTCCAAAATTCGGCCCTCGATCACAAGGATATGATATATTTCGTAAACGAATTCGAAAGGCTCGGCAGAGACATATTAGTCTAA
Protein
MPADSDLIVAEREDLGIDRTLYKSLSERSKHEDFIRRAVDLLVERVVFGRSLRTSKVVEWAVPSEIKKAIDLKPRDGPISHDELLGLMADVARYSVNTAHPYFVNQLYSSVDPYGLVGQWLTDALNPSVYTYEVAPVFTLMEEEVLKEMRVLVGWKDGEGDGIFCPGGSIANGYAISCARFHFYPEIKTKGVYAVPKLTLYTSELAHYSTKKLAAFMGIGDENCVLIKTDKYGKIDVEDLEAKIVEGIEEGAAPFLVTATAGTTVFGAFDPLVPIAALCKKYNLWLHVDAAWGGGALMSKKHRHLLNGIELADSVTWNPHKLLAAPQQCSTFLIRHKNVLKEGHSCNAKYLFQKDKFYDTSYDTGDKHIQCGRRADVLKFWFMWKAKGSDGFENHIDTLFDNARFFLEQIRNREGFELVIEKPECTNIMFWYVPRCLRGCENESDYRERLHKVAPKIKELMIKEGSMMVTYQPQGDLVNFFRIVFQNSALDHKDMIYFVNEFERLGRDILV
Summary
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the group II decarboxylase family.
Uniprot
H9JRC8
A0A0C5QEV9
A0A0L7LFT8
A0A2H1WRM9
A0A194PEG0
D4AH66
+ More
A0A194R3E1 A0A3S2TSR1 A0A0G3VGT7 A0A2A4JAM0 G8FGR7 G4XH89 A0A212EPV4 A0A2J7PB96 A0A067QKQ9 A9YVA8 A7U8C7 A0A1B6GV38 A0A2H8TVD8 A0A182XNP4 A0A182LMV3 A0A182I7A3 Q7PWN7 A0A182UR97 A0A182QH49 J9KAH8 A0A182P8A6 A0A182Y0A8 A0A182J8I3 A0A182LVI3 A0A182VUD2 A0A182RSJ2 A0A084VL13 A0A182MZ80 A0A1S4FGS8 Q171S0 A0A336MJ76 A0A2S2Q2F2 A0A1Y1MWE9 A0A0T6BCB4 A0A1W4WSF2 A0A1Q3FT64 A0A1Q3FT96 U4UTD4 A0A154PAD6 N6TKV9 B0WRQ9 A0A0L0CK42 B4JCX3 K7IY18 A0A3B0K3M2 A0A0J9R2X0 B3MN27 B4HXA1 A0A1Q3FSS4 A0A1A9YJW1 A0A0A9XUT1 B4Q567 A0A0L7RAZ6 B4KIX9 A0A1W4V741 A0A1B0BT94 A0A023F549 A0A0M4E5T9 A0A3B0KBV2 A0A1J1HN78 Q24062 A0A1L8DZC2 A0A1I8Q1Y9 A0A1A9UYR8 B4GX04 B4P3Q4 A0A0N0BFM6 B4N0X5 X2J8B3 B3NAD2 B4M8L3 Q29K73 A0A087ZNY3 A0A1A9Z445 T1I1H2 A0A1I8ME86 A0A2A3E1X6 W8AVS4 A0A1S3D4P7 A0A1A9WXD3 A0A0C9R8F6 A0A0K8VN41 A0A1B0G692 A0A034WAD5 A0A182F2K1 A0A232FDU5 A0A0A1X1S4 A0A195CYF4 A0A026WF04 A0A151JW08 E1ZV07 A0A3L8DKF1 A0A151JQB3 F4WHT2
A0A194R3E1 A0A3S2TSR1 A0A0G3VGT7 A0A2A4JAM0 G8FGR7 G4XH89 A0A212EPV4 A0A2J7PB96 A0A067QKQ9 A9YVA8 A7U8C7 A0A1B6GV38 A0A2H8TVD8 A0A182XNP4 A0A182LMV3 A0A182I7A3 Q7PWN7 A0A182UR97 A0A182QH49 J9KAH8 A0A182P8A6 A0A182Y0A8 A0A182J8I3 A0A182LVI3 A0A182VUD2 A0A182RSJ2 A0A084VL13 A0A182MZ80 A0A1S4FGS8 Q171S0 A0A336MJ76 A0A2S2Q2F2 A0A1Y1MWE9 A0A0T6BCB4 A0A1W4WSF2 A0A1Q3FT64 A0A1Q3FT96 U4UTD4 A0A154PAD6 N6TKV9 B0WRQ9 A0A0L0CK42 B4JCX3 K7IY18 A0A3B0K3M2 A0A0J9R2X0 B3MN27 B4HXA1 A0A1Q3FSS4 A0A1A9YJW1 A0A0A9XUT1 B4Q567 A0A0L7RAZ6 B4KIX9 A0A1W4V741 A0A1B0BT94 A0A023F549 A0A0M4E5T9 A0A3B0KBV2 A0A1J1HN78 Q24062 A0A1L8DZC2 A0A1I8Q1Y9 A0A1A9UYR8 B4GX04 B4P3Q4 A0A0N0BFM6 B4N0X5 X2J8B3 B3NAD2 B4M8L3 Q29K73 A0A087ZNY3 A0A1A9Z445 T1I1H2 A0A1I8ME86 A0A2A3E1X6 W8AVS4 A0A1S3D4P7 A0A1A9WXD3 A0A0C9R8F6 A0A0K8VN41 A0A1B0G692 A0A034WAD5 A0A182F2K1 A0A232FDU5 A0A0A1X1S4 A0A195CYF4 A0A026WF04 A0A151JW08 E1ZV07 A0A3L8DKF1 A0A151JQB3 F4WHT2
Pubmed
19121390
26227816
26354079
22234245
22118469
24845553
+ More
19366687 20966253 12364791 14747013 17210077 25244985 24438588 17510324 28004739 23537049 26108605 17994087 20075255 22936249 25401762 25474469 8376987 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 25315136 24495485 25348373 28648823 25830018 24508170 20798317 30249741 21719571
19366687 20966253 12364791 14747013 17210077 25244985 24438588 17510324 28004739 23537049 26108605 17994087 20075255 22936249 25401762 25474469 8376987 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 25315136 24495485 25348373 28648823 25830018 24508170 20798317 30249741 21719571
EMBL
BABH01016128
KM523624
AJQ30182.1
KM523625
AJQ30183.1
JTDY01001309
+ More
KOB74239.1 ODYU01010161 SOQ55084.1 KQ459606 KPI91741.1 AB503723 BAI87832.1 KQ460779 KPJ12323.1 RSAL01000007 RVE54119.1 KP657641 AKL78866.1 NWSH01002373 PCG68443.1 JN003850 AEQ77286.1 JF811444 AEP43793.1 AGBW02013361 OWR43523.1 NEVH01027131 PNF13607.1 KK853239 KDR09499.1 EU302526 ABX89951.1 EU019709 ABU25221.1 GECZ01003514 JAS66255.1 GFXV01006382 MBW18187.1 APCN01003374 AAAB01008984 EAA14875.5 AXCN02002069 ABLF02018654 AXCM01000345 ATLV01014395 KE524970 KFB38657.1 CH477447 EAT40747.1 UFQS01001093 UFQT01001093 SSX08972.1 SSX28883.1 GGMS01002618 MBY71821.1 GEZM01020016 JAV89458.1 LJIG01001997 KRT84943.1 GFDL01004403 JAV30642.1 GFDL01004402 JAV30643.1 KB632355 ERL93380.1 KQ434857 KZC08803.1 APGK01033004 APGK01033005 KB740860 ENN78563.1 DS232058 EDS33463.1 JRES01000300 KNC32620.1 CH916368 EDW03212.1 OUUW01000006 SPP82540.1 CM002910 KMY90164.1 CH902620 EDV32005.1 CH480818 EDW51681.1 GFDL01004401 JAV30644.1 GBHO01019032 GBRD01000590 JAG24572.1 JAG65231.1 CM000361 EDX04973.1 KQ414618 KOC67911.1 CH933807 EDW13492.1 JXJN01020065 GBBI01002528 JAC16184.1 CP012523 ALC39546.1 SPP82541.1 CVRI01000010 CRK88836.1 U01239 AE014134 AY069752 AAC46466.1 AAF53337.1 AAL39897.1 AFH03699.1 GFDF01002254 JAV11830.1 CH479195 EDW27331.1 CM000157 EDW88355.1 KQ435798 KOX73314.1 CH963920 EDW77738.1 AHN54424.1 CH954177 EDV58634.1 CH940654 EDW57539.1 CH379061 EAL33304.2 ACPB03006877 KZ288443 PBC25695.1 GAMC01017728 GAMC01017727 GAMC01017726 GAMC01017725 GAMC01017724 GAMC01017723 JAB88828.1 GBYB01008619 GBYB01009157 JAG78386.1 JAG78924.1 GDHF01012017 JAI40297.1 CCAG010006778 GAKP01007298 JAC51654.1 NNAY01000405 OXU28629.1 GBXI01009220 JAD05072.1 KQ977171 KYN05174.1 KK107238 EZA54612.1 KQ981688 KYN37926.1 GL434335 EFN75005.1 QOIP01000007 RLU20663.1 KQ978684 KYN29300.1 GL888167 EGI66289.1
KOB74239.1 ODYU01010161 SOQ55084.1 KQ459606 KPI91741.1 AB503723 BAI87832.1 KQ460779 KPJ12323.1 RSAL01000007 RVE54119.1 KP657641 AKL78866.1 NWSH01002373 PCG68443.1 JN003850 AEQ77286.1 JF811444 AEP43793.1 AGBW02013361 OWR43523.1 NEVH01027131 PNF13607.1 KK853239 KDR09499.1 EU302526 ABX89951.1 EU019709 ABU25221.1 GECZ01003514 JAS66255.1 GFXV01006382 MBW18187.1 APCN01003374 AAAB01008984 EAA14875.5 AXCN02002069 ABLF02018654 AXCM01000345 ATLV01014395 KE524970 KFB38657.1 CH477447 EAT40747.1 UFQS01001093 UFQT01001093 SSX08972.1 SSX28883.1 GGMS01002618 MBY71821.1 GEZM01020016 JAV89458.1 LJIG01001997 KRT84943.1 GFDL01004403 JAV30642.1 GFDL01004402 JAV30643.1 KB632355 ERL93380.1 KQ434857 KZC08803.1 APGK01033004 APGK01033005 KB740860 ENN78563.1 DS232058 EDS33463.1 JRES01000300 KNC32620.1 CH916368 EDW03212.1 OUUW01000006 SPP82540.1 CM002910 KMY90164.1 CH902620 EDV32005.1 CH480818 EDW51681.1 GFDL01004401 JAV30644.1 GBHO01019032 GBRD01000590 JAG24572.1 JAG65231.1 CM000361 EDX04973.1 KQ414618 KOC67911.1 CH933807 EDW13492.1 JXJN01020065 GBBI01002528 JAC16184.1 CP012523 ALC39546.1 SPP82541.1 CVRI01000010 CRK88836.1 U01239 AE014134 AY069752 AAC46466.1 AAF53337.1 AAL39897.1 AFH03699.1 GFDF01002254 JAV11830.1 CH479195 EDW27331.1 CM000157 EDW88355.1 KQ435798 KOX73314.1 CH963920 EDW77738.1 AHN54424.1 CH954177 EDV58634.1 CH940654 EDW57539.1 CH379061 EAL33304.2 ACPB03006877 KZ288443 PBC25695.1 GAMC01017728 GAMC01017727 GAMC01017726 GAMC01017725 GAMC01017724 GAMC01017723 JAB88828.1 GBYB01008619 GBYB01009157 JAG78386.1 JAG78924.1 GDHF01012017 JAI40297.1 CCAG010006778 GAKP01007298 JAC51654.1 NNAY01000405 OXU28629.1 GBXI01009220 JAD05072.1 KQ977171 KYN05174.1 KK107238 EZA54612.1 KQ981688 KYN37926.1 GL434335 EFN75005.1 QOIP01000007 RLU20663.1 KQ978684 KYN29300.1 GL888167 EGI66289.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000283053
UP000218220
+ More
UP000007151 UP000235965 UP000027135 UP000076407 UP000075882 UP000075840 UP000007062 UP000075903 UP000075886 UP000007819 UP000075885 UP000076408 UP000075880 UP000075883 UP000075920 UP000075900 UP000030765 UP000075884 UP000008820 UP000192223 UP000030742 UP000076502 UP000019118 UP000002320 UP000037069 UP000001070 UP000002358 UP000268350 UP000007801 UP000001292 UP000092443 UP000000304 UP000053825 UP000009192 UP000192221 UP000092460 UP000092553 UP000183832 UP000000803 UP000095300 UP000078200 UP000008744 UP000002282 UP000053105 UP000007798 UP000008711 UP000008792 UP000001819 UP000005203 UP000092445 UP000015103 UP000095301 UP000242457 UP000079169 UP000091820 UP000092444 UP000069272 UP000215335 UP000078542 UP000053097 UP000078541 UP000000311 UP000279307 UP000078492 UP000007755
UP000007151 UP000235965 UP000027135 UP000076407 UP000075882 UP000075840 UP000007062 UP000075903 UP000075886 UP000007819 UP000075885 UP000076408 UP000075880 UP000075883 UP000075920 UP000075900 UP000030765 UP000075884 UP000008820 UP000192223 UP000030742 UP000076502 UP000019118 UP000002320 UP000037069 UP000001070 UP000002358 UP000268350 UP000007801 UP000001292 UP000092443 UP000000304 UP000053825 UP000009192 UP000192221 UP000092460 UP000092553 UP000183832 UP000000803 UP000095300 UP000078200 UP000008744 UP000002282 UP000053105 UP000007798 UP000008711 UP000008792 UP000001819 UP000005203 UP000092445 UP000015103 UP000095301 UP000242457 UP000079169 UP000091820 UP000092444 UP000069272 UP000215335 UP000078542 UP000053097 UP000078541 UP000000311 UP000279307 UP000078492 UP000007755
Pfam
Interpro
IPR015424
PyrdxlP-dep_Trfase
+ More
IPR002129 PyrdxlP-dep_de-COase
IPR015421 PyrdxlP-dep_Trfase_major
IPR001895 RASGEF_cat_dom
IPR035899 DBL_dom_sf
IPR002119 Histone_H2A
IPR008937 Ras-like_GEF
IPR023578 Ras_GEF_dom_sf
IPR001849 PH_domain
IPR000219 DH-domain
IPR009072 Histone-fold
IPR019804 Ras_G-nucl-exch_fac_CS
IPR007125 Histone_H2A/H2B/H3
IPR011993 PH-like_dom_sf
IPR000651 Ras-like_Gua-exchang_fac_N
IPR032629 DCB_dom
IPR015403 Sec7_C
IPR000904 Sec7_dom
IPR016024 ARM-type_fold
IPR032691 Sec7_N
IPR035999 Sec7_dom_sf
IPR023394 Sec7_C_sf
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR002129 PyrdxlP-dep_de-COase
IPR015421 PyrdxlP-dep_Trfase_major
IPR001895 RASGEF_cat_dom
IPR035899 DBL_dom_sf
IPR002119 Histone_H2A
IPR008937 Ras-like_GEF
IPR023578 Ras_GEF_dom_sf
IPR001849 PH_domain
IPR000219 DH-domain
IPR009072 Histone-fold
IPR019804 Ras_G-nucl-exch_fac_CS
IPR007125 Histone_H2A/H2B/H3
IPR011993 PH-like_dom_sf
IPR000651 Ras-like_Gua-exchang_fac_N
IPR032629 DCB_dom
IPR015403 Sec7_C
IPR000904 Sec7_dom
IPR016024 ARM-type_fold
IPR032691 Sec7_N
IPR035999 Sec7_dom_sf
IPR023394 Sec7_C_sf
IPR015422 PyrdxlP-dep_Trfase_dom1
SUPFAM
ProteinModelPortal
H9JRC8
A0A0C5QEV9
A0A0L7LFT8
A0A2H1WRM9
A0A194PEG0
D4AH66
+ More
A0A194R3E1 A0A3S2TSR1 A0A0G3VGT7 A0A2A4JAM0 G8FGR7 G4XH89 A0A212EPV4 A0A2J7PB96 A0A067QKQ9 A9YVA8 A7U8C7 A0A1B6GV38 A0A2H8TVD8 A0A182XNP4 A0A182LMV3 A0A182I7A3 Q7PWN7 A0A182UR97 A0A182QH49 J9KAH8 A0A182P8A6 A0A182Y0A8 A0A182J8I3 A0A182LVI3 A0A182VUD2 A0A182RSJ2 A0A084VL13 A0A182MZ80 A0A1S4FGS8 Q171S0 A0A336MJ76 A0A2S2Q2F2 A0A1Y1MWE9 A0A0T6BCB4 A0A1W4WSF2 A0A1Q3FT64 A0A1Q3FT96 U4UTD4 A0A154PAD6 N6TKV9 B0WRQ9 A0A0L0CK42 B4JCX3 K7IY18 A0A3B0K3M2 A0A0J9R2X0 B3MN27 B4HXA1 A0A1Q3FSS4 A0A1A9YJW1 A0A0A9XUT1 B4Q567 A0A0L7RAZ6 B4KIX9 A0A1W4V741 A0A1B0BT94 A0A023F549 A0A0M4E5T9 A0A3B0KBV2 A0A1J1HN78 Q24062 A0A1L8DZC2 A0A1I8Q1Y9 A0A1A9UYR8 B4GX04 B4P3Q4 A0A0N0BFM6 B4N0X5 X2J8B3 B3NAD2 B4M8L3 Q29K73 A0A087ZNY3 A0A1A9Z445 T1I1H2 A0A1I8ME86 A0A2A3E1X6 W8AVS4 A0A1S3D4P7 A0A1A9WXD3 A0A0C9R8F6 A0A0K8VN41 A0A1B0G692 A0A034WAD5 A0A182F2K1 A0A232FDU5 A0A0A1X1S4 A0A195CYF4 A0A026WF04 A0A151JW08 E1ZV07 A0A3L8DKF1 A0A151JQB3 F4WHT2
A0A194R3E1 A0A3S2TSR1 A0A0G3VGT7 A0A2A4JAM0 G8FGR7 G4XH89 A0A212EPV4 A0A2J7PB96 A0A067QKQ9 A9YVA8 A7U8C7 A0A1B6GV38 A0A2H8TVD8 A0A182XNP4 A0A182LMV3 A0A182I7A3 Q7PWN7 A0A182UR97 A0A182QH49 J9KAH8 A0A182P8A6 A0A182Y0A8 A0A182J8I3 A0A182LVI3 A0A182VUD2 A0A182RSJ2 A0A084VL13 A0A182MZ80 A0A1S4FGS8 Q171S0 A0A336MJ76 A0A2S2Q2F2 A0A1Y1MWE9 A0A0T6BCB4 A0A1W4WSF2 A0A1Q3FT64 A0A1Q3FT96 U4UTD4 A0A154PAD6 N6TKV9 B0WRQ9 A0A0L0CK42 B4JCX3 K7IY18 A0A3B0K3M2 A0A0J9R2X0 B3MN27 B4HXA1 A0A1Q3FSS4 A0A1A9YJW1 A0A0A9XUT1 B4Q567 A0A0L7RAZ6 B4KIX9 A0A1W4V741 A0A1B0BT94 A0A023F549 A0A0M4E5T9 A0A3B0KBV2 A0A1J1HN78 Q24062 A0A1L8DZC2 A0A1I8Q1Y9 A0A1A9UYR8 B4GX04 B4P3Q4 A0A0N0BFM6 B4N0X5 X2J8B3 B3NAD2 B4M8L3 Q29K73 A0A087ZNY3 A0A1A9Z445 T1I1H2 A0A1I8ME86 A0A2A3E1X6 W8AVS4 A0A1S3D4P7 A0A1A9WXD3 A0A0C9R8F6 A0A0K8VN41 A0A1B0G692 A0A034WAD5 A0A182F2K1 A0A232FDU5 A0A0A1X1S4 A0A195CYF4 A0A026WF04 A0A151JW08 E1ZV07 A0A3L8DKF1 A0A151JQB3 F4WHT2
PDB
2JIS
E-value=2.25691e-147,
Score=1340
Ontologies
PATHWAY
00250
Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00430 Taurine and hypotaurine metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00430 Taurine and hypotaurine metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
PANTHER
Topology
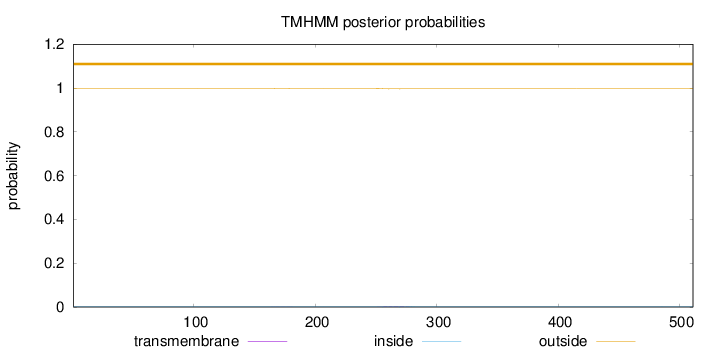
Length:
511
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07045
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00124
outside
1 - 511
Population Genetic Test Statistics
Pi
4.098893
Theta
14.40652
Tajima's D
-1.79803
CLR
199.943654
CSRT
0.0256987150642468
Interpretation
Uncertain
