Gene
KWMTBOMO06854
Pre Gene Modal
BGIBMGA012068
Annotation
PREDICTED:_multiple_C2_and_transmembrane_domain-containing_protein_1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.27 Nuclear Reliability : 1.82 PlasmaMembrane Reliability : 1.292
Sequence
CDS
ATGGGTGAAAGAGCTAAAAGTCACTTCGGTGATTTTAAGAAGACACATTTCGCCAAACTGCACGAGAGGATACAAAGGAAATATGAAGAGATGCAATGGAAACTGGAGAAATCAAAATCCGTTGACGTACTGTCGACGCTTAAAGATGATTATGCCTTTAAAGAGATCAAATTCGCAAGCACTTTGGATATAACTGCGGAGTCGGACGAAATTTTTGAAGAAGTTCAGAACCATAGTTCCTTGGAAAAGCATACATTGCTAATTAAAGACATGAATAGGAATGACGCTAATCTTGAATATGTTGATTTCAAAGAAGCTAGTATTTCCGAGTCCGAAGGGTTTACTCCTCTTGAAAAGTACGAAAATGGCAGGGAAGATAAATTCTTTGGAGATGACTTTTCTAAAATGAGTCTTGAGTCATTAAATGAAGTAGCTTCAGGGAGCGACGGTGAGCTCACGCCCCCGGCGACTCCCCCGAAGATTTCGATTCGCAATAGAATTGGTTCAAAAATATCCGCTGTCAAGGAAAAACGTTGGCGAGAGGAAAAAGTAAAAAATCCGAACAAGGACAGGAAAAGGGAACATCGTGACAAAGTTGAAAAGTTTATTTTGAGTAATACGACTAAACAGGATTTGAGGAGCTTAAAGAAGACTAAATTGGGTACTGTAACAATTGCACTGATCGACGCTACAGATTTAGAGACTGAAGGCCCCGAAGAAAGAACCCGTATGTTATTTTGTCGATTTAGATTGGGAACTGAGAAACATAAGTCAAAGACAGCAAAAGGGAATACAACTAAATTGAAATGGCAAGAACTATTCAGTTTGAGTATGTACGAAGACCATTTGCTTGAGTTGTCTCTGTGCGATAAAGATTTCTGTATTGGCAGGACTATAATAGATCTGAATAATTTGGATAAAGAGAAGACACATGAAATGCGTCTCAAGATAGAAGGGGAAATATCCGTAGTACAAATTTTTCTATTAATTACAATCAGCGGTATACCGTTACACAATACCATCGCAGATTTAGAAGATTATGAAGCAACAACCAAGGAGCTCGATGTTGTCAAAAAGAAATTTGCATGGTACCATATCGGGAATTCAAACGAAGTTGGCTGGTTATCTGTGTTGGTATACGGAGCTAAAGGATTACATGCTCAGGACACGTGTTGCATATTAGAACTTGATAATGAAAGGTTACAAACACATACAGAAATCAAAACGAATGAACCTAATTGGATGAAATTATTTAGATTCATTGTAAATGATATTACTTCAGTAATAGAAGTGACAGTACATGACGAAAAAAGATCTGAGGATATTGCAAAAATTACAATACCATTATTAAAAATTAACAACGGAGTAAAAAAATGGTATGCTCTGAAACACACAACGCAGAGAGAAAGAGCGAGAGGAACGAACCCTAGAATCCTGTTAGAAATGAAAGTCACATGGAATCTCACAAAAGCATCGCTCCGATCGTTTACACCCAAACAGACAAAATATTTAGAAACGAAAGCCAAATTCGATAGACATATTTTTGCGAGGAATATCTCAAGGGCAAAATATGTCACTGCATGGGTGCTAAATGCTATCCAGATCGTTAAAACATGTTTCGAATGGGAATCAAGAAAATCAAACGCATTAGCCCTCACATTATGGCTATTATTTTGTTGGTTTTTCAAAATGTGGATGTTGCCACTTCTATTATTGATACCATTCATTTGGTATCGTCCACCGAGATACTATTTAGTAAACTGGAAACGTTATTTCAAAGGAATGCCAATTGAAGATGATAATTCCAAGAAAGAGAAGGACGAAAAAAACACATCTTTGAGACAAAAGATACAAAGTTTACAGGAAATGGTACAGACGGTGCAGAATTTCATTGGAACGTTTGCGAGTCTACACGAGAGAGTAAAGAATTTGTTCAACTTTACAGTACCCTTCCTAAGTTTCTTGACCATATTCATGATCATAATTATCGCCTTCGTAATGTTTGTGATTCCTGTGAAATATATCTTCATGGGTTGGGGTGTACACAAATTCACAAGAAAAATTCTTAGGCCGAACAGAATTCCAAATAATGAAATTCTGGATCTGCTCTCCCGAGTACCTGATGATGAAACGTTGCTTGACTGTGAAGAGCTACCTTTGGAGGAACCAGAGGAAGACGTAGTAGAACCATAA
Protein
MGERAKSHFGDFKKTHFAKLHERIQRKYEEMQWKLEKSKSVDVLSTLKDDYAFKEIKFASTLDITAESDEIFEEVQNHSSLEKHTLLIKDMNRNDANLEYVDFKEASISESEGFTPLEKYENGREDKFFGDDFSKMSLESLNEVASGSDGELTPPATPPKISIRNRIGSKISAVKEKRWREEKVKNPNKDRKREHRDKVEKFILSNTTKQDLRSLKKTKLGTVTIALIDATDLETEGPEERTRMLFCRFRLGTEKHKSKTAKGNTTKLKWQELFSLSMYEDHLLELSLCDKDFCIGRTIIDLNNLDKEKTHEMRLKIEGEISVVQIFLLITISGIPLHNTIADLEDYEATTKELDVVKKKFAWYHIGNSNEVGWLSVLVYGAKGLHAQDTCCILELDNERLQTHTEIKTNEPNWMKLFRFIVNDITSVIEVTVHDEKRSEDIAKITIPLLKINNGVKKWYALKHTTQRERARGTNPRILLEMKVTWNLTKASLRSFTPKQTKYLETKAKFDRHIFARNISRAKYVTAWVLNAIQIVKTCFEWESRKSNALALTLWLLFCWFFKMWMLPLLLLIPFIWYRPPRYYLVNWKRYFKGMPIEDDNSKKEKDEKNTSLRQKIQSLQEMVQTVQNFIGTFASLHERVKNLFNFTVPFLSFLTIFMIIIIAFVMFVIPVKYIFMGWGVHKFTRKILRPNRIPNNEILDLLSRVPDDETLLDCEELPLEEPEEDVVEP
Summary
Uniprot
A0A2A4JA84
A0A2H1WPT5
A0A3S2M9K5
A0A1E1WBJ9
A0A194R477
A0A212EPY1
+ More
A0A194PEP1 A0A0L7LFK5 A0A2H1WBS6 H9JW63 A0A2W1BKK6 A0A2A4K8X0 A0A2A4K984 A0A1B6EUE0 A0A194R7I2 A0A3S2NMQ7 A0A1Y1M3S5 A0A2A3EPA0 A0A067QZE1 V9IEV3 V9IFE7 A0A310S7H1 E0VQX2 A0A158NNP2 A0A1Y1M0D1 A0A0C9RHL5 A0A232FFH3 A0A151I9P9 T1I1Y3 A0A2A4J601 A0A2A4J4U8 A0A1Y1M5V4 D6WUE3 A0A139WFX4 A0A139WFN8 A0A088A9U8 A0A1I8PKP5 A0A0M9A7A8 A0A154NZ39 A0A151X947 A0A212F6Q9 A0A195F6N0 A0A1I8MDU5 U4UKD4 A0A2H1V383 E2AYX5 E9GTE2 N6TJP3 A0A0P5PPJ2 A0A0P6E172 A0A0P5G832 A0A0P5IUY0 A0A0N8C642 A0A0P5EK68 A0A0P5HKE5 A0A0P5HJB2 A0A0P5Q3V5 A0A0P5RTR2 A0A1A9URC3 A0A0N8AJP7 A0A0P5JBY1 A0A0P5HPH7 F4WQE3 A0A0P5I1X2 A0A195B3G7
A0A194PEP1 A0A0L7LFK5 A0A2H1WBS6 H9JW63 A0A2W1BKK6 A0A2A4K8X0 A0A2A4K984 A0A1B6EUE0 A0A194R7I2 A0A3S2NMQ7 A0A1Y1M3S5 A0A2A3EPA0 A0A067QZE1 V9IEV3 V9IFE7 A0A310S7H1 E0VQX2 A0A158NNP2 A0A1Y1M0D1 A0A0C9RHL5 A0A232FFH3 A0A151I9P9 T1I1Y3 A0A2A4J601 A0A2A4J4U8 A0A1Y1M5V4 D6WUE3 A0A139WFX4 A0A139WFN8 A0A088A9U8 A0A1I8PKP5 A0A0M9A7A8 A0A154NZ39 A0A151X947 A0A212F6Q9 A0A195F6N0 A0A1I8MDU5 U4UKD4 A0A2H1V383 E2AYX5 E9GTE2 N6TJP3 A0A0P5PPJ2 A0A0P6E172 A0A0P5G832 A0A0P5IUY0 A0A0N8C642 A0A0P5EK68 A0A0P5HKE5 A0A0P5HJB2 A0A0P5Q3V5 A0A0P5RTR2 A0A1A9URC3 A0A0N8AJP7 A0A0P5JBY1 A0A0P5HPH7 F4WQE3 A0A0P5I1X2 A0A195B3G7
Pubmed
EMBL
NWSH01002373
PCG68444.1
ODYU01010161
SOQ55085.1
RSAL01000007
RVE54120.1
+ More
GDQN01006737 JAT84317.1 KQ460779 KPJ12324.1 AGBW02013361 OWR43524.1 KQ459606 KPI91742.1 JTDY01001309 KOB74242.1 ODYU01007596 SOQ50513.1 BABH01025337 BABH01025338 KZ150109 PZC73376.1 NWSH01000040 PCG80354.1 PCG80353.1 GECZ01028281 JAS41488.1 KQ460615 KPJ13607.1 RSAL01000237 RVE43708.1 GEZM01043132 JAV79290.1 KZ288212 PBC32861.1 KK852859 KDR14857.1 JR040572 AEY59192.1 JR040571 AEY59191.1 KQ773232 OAD52398.1 DS235442 EEB15778.1 ADTU01021618 ADTU01021619 GEZM01043131 JAV79292.1 GBYB01000914 GBYB01006426 JAG70681.1 JAG76193.1 NNAY01000316 OXU29270.1 KQ978273 KYM95768.1 ACPB03005944 NWSH01002972 PCG67166.1 PCG67167.1 GEZM01043135 JAV79286.1 KQ971352 EFA07297.2 KYB26751.1 KYB26752.1 KQ435732 KOX77403.1 KQ434783 KZC04883.1 KQ982383 KYQ56897.1 AGBW02009983 OWR49417.1 KQ981744 KYN36255.1 KB632339 ERL92928.1 ODYU01000314 SOQ34832.1 GL444028 EFN61345.1 GL732563 EFX77264.1 APGK01034847 APGK01034848 APGK01034849 APGK01034850 KB740923 ENN78073.1 GDIQ01131074 JAL20652.1 GDIQ01073928 JAN20809.1 GDIQ01244941 JAK06784.1 GDIQ01216234 JAK35491.1 GDIQ01111123 JAL40603.1 GDIQ01269085 JAJ82639.1 GDIQ01226131 GDIQ01226129 JAK25596.1 GDIQ01226130 JAK25595.1 GDIQ01119462 JAL32264.1 GDIQ01196991 GDIQ01097322 JAL54404.1 GDIQ01269083 JAJ82641.1 GDIQ01207896 JAK43829.1 GDIQ01224927 GDIQ01224925 JAK26798.1 GL888273 EGI63543.1 GDIQ01224926 JAK26799.1 KQ976641 KYM78739.1
GDQN01006737 JAT84317.1 KQ460779 KPJ12324.1 AGBW02013361 OWR43524.1 KQ459606 KPI91742.1 JTDY01001309 KOB74242.1 ODYU01007596 SOQ50513.1 BABH01025337 BABH01025338 KZ150109 PZC73376.1 NWSH01000040 PCG80354.1 PCG80353.1 GECZ01028281 JAS41488.1 KQ460615 KPJ13607.1 RSAL01000237 RVE43708.1 GEZM01043132 JAV79290.1 KZ288212 PBC32861.1 KK852859 KDR14857.1 JR040572 AEY59192.1 JR040571 AEY59191.1 KQ773232 OAD52398.1 DS235442 EEB15778.1 ADTU01021618 ADTU01021619 GEZM01043131 JAV79292.1 GBYB01000914 GBYB01006426 JAG70681.1 JAG76193.1 NNAY01000316 OXU29270.1 KQ978273 KYM95768.1 ACPB03005944 NWSH01002972 PCG67166.1 PCG67167.1 GEZM01043135 JAV79286.1 KQ971352 EFA07297.2 KYB26751.1 KYB26752.1 KQ435732 KOX77403.1 KQ434783 KZC04883.1 KQ982383 KYQ56897.1 AGBW02009983 OWR49417.1 KQ981744 KYN36255.1 KB632339 ERL92928.1 ODYU01000314 SOQ34832.1 GL444028 EFN61345.1 GL732563 EFX77264.1 APGK01034847 APGK01034848 APGK01034849 APGK01034850 KB740923 ENN78073.1 GDIQ01131074 JAL20652.1 GDIQ01073928 JAN20809.1 GDIQ01244941 JAK06784.1 GDIQ01216234 JAK35491.1 GDIQ01111123 JAL40603.1 GDIQ01269085 JAJ82639.1 GDIQ01226131 GDIQ01226129 JAK25596.1 GDIQ01226130 JAK25595.1 GDIQ01119462 JAL32264.1 GDIQ01196991 GDIQ01097322 JAL54404.1 GDIQ01269083 JAJ82641.1 GDIQ01207896 JAK43829.1 GDIQ01224927 GDIQ01224925 JAK26798.1 GL888273 EGI63543.1 GDIQ01224926 JAK26799.1 KQ976641 KYM78739.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000005204 UP000242457 UP000027135 UP000009046 UP000005205 UP000215335 UP000078542 UP000015103 UP000007266 UP000005203 UP000095300 UP000053105 UP000076502 UP000075809 UP000078541 UP000095301 UP000030742 UP000000311 UP000000305 UP000019118 UP000078200 UP000007755 UP000078540
UP000005204 UP000242457 UP000027135 UP000009046 UP000005205 UP000215335 UP000078542 UP000015103 UP000007266 UP000005203 UP000095300 UP000053105 UP000076502 UP000075809 UP000078541 UP000095301 UP000030742 UP000000311 UP000000305 UP000019118 UP000078200 UP000007755 UP000078540
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JA84
A0A2H1WPT5
A0A3S2M9K5
A0A1E1WBJ9
A0A194R477
A0A212EPY1
+ More
A0A194PEP1 A0A0L7LFK5 A0A2H1WBS6 H9JW63 A0A2W1BKK6 A0A2A4K8X0 A0A2A4K984 A0A1B6EUE0 A0A194R7I2 A0A3S2NMQ7 A0A1Y1M3S5 A0A2A3EPA0 A0A067QZE1 V9IEV3 V9IFE7 A0A310S7H1 E0VQX2 A0A158NNP2 A0A1Y1M0D1 A0A0C9RHL5 A0A232FFH3 A0A151I9P9 T1I1Y3 A0A2A4J601 A0A2A4J4U8 A0A1Y1M5V4 D6WUE3 A0A139WFX4 A0A139WFN8 A0A088A9U8 A0A1I8PKP5 A0A0M9A7A8 A0A154NZ39 A0A151X947 A0A212F6Q9 A0A195F6N0 A0A1I8MDU5 U4UKD4 A0A2H1V383 E2AYX5 E9GTE2 N6TJP3 A0A0P5PPJ2 A0A0P6E172 A0A0P5G832 A0A0P5IUY0 A0A0N8C642 A0A0P5EK68 A0A0P5HKE5 A0A0P5HJB2 A0A0P5Q3V5 A0A0P5RTR2 A0A1A9URC3 A0A0N8AJP7 A0A0P5JBY1 A0A0P5HPH7 F4WQE3 A0A0P5I1X2 A0A195B3G7
A0A194PEP1 A0A0L7LFK5 A0A2H1WBS6 H9JW63 A0A2W1BKK6 A0A2A4K8X0 A0A2A4K984 A0A1B6EUE0 A0A194R7I2 A0A3S2NMQ7 A0A1Y1M3S5 A0A2A3EPA0 A0A067QZE1 V9IEV3 V9IFE7 A0A310S7H1 E0VQX2 A0A158NNP2 A0A1Y1M0D1 A0A0C9RHL5 A0A232FFH3 A0A151I9P9 T1I1Y3 A0A2A4J601 A0A2A4J4U8 A0A1Y1M5V4 D6WUE3 A0A139WFX4 A0A139WFN8 A0A088A9U8 A0A1I8PKP5 A0A0M9A7A8 A0A154NZ39 A0A151X947 A0A212F6Q9 A0A195F6N0 A0A1I8MDU5 U4UKD4 A0A2H1V383 E2AYX5 E9GTE2 N6TJP3 A0A0P5PPJ2 A0A0P6E172 A0A0P5G832 A0A0P5IUY0 A0A0N8C642 A0A0P5EK68 A0A0P5HKE5 A0A0P5HJB2 A0A0P5Q3V5 A0A0P5RTR2 A0A1A9URC3 A0A0N8AJP7 A0A0P5JBY1 A0A0P5HPH7 F4WQE3 A0A0P5I1X2 A0A195B3G7
PDB
2EP6
E-value=1.78546e-15,
Score=204
Ontologies
KEGG
GO
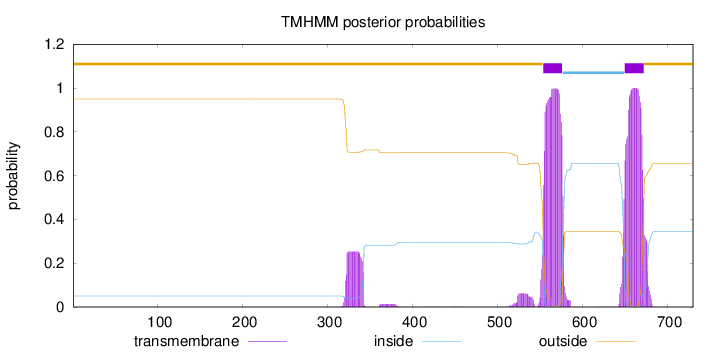
Topology
Length:
730
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
53.24711
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05023
outside
1 - 553
TMhelix
554 - 576
inside
577 - 649
TMhelix
650 - 672
outside
673 - 730
Population Genetic Test Statistics
Pi
3.634786
Theta
4.319301
Tajima's D
-1.853958
CLR
345.012943
CSRT
0.0190990450477476
Interpretation
Possibly Positive selection
