Gene
KWMTBOMO06844
Annotation
PREDICTED:_uncharacterized_protein_LOC106129555_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.487
Sequence
CDS
ATGAAGTGCGCCGGTTGTCTAAAGGTAACGAAAGAATTAGGTTCTATTCGTTGTACCAGCTCATCTTGTACGAAAATTTTCTGTCCTCTCTGTATAGACATCTCTCTGGTTCCTGTAGATCGAAAAAAGATTTGGAAGTGTCCTGATTGTTGTGCTGCACACAAAAAAGGAGGTGATAACAGTCTTACTCCTATAAGAGGTACATATGACGGCCAAAATATAACGACTCGTAAGAAGGAGCCCACTGCAGAGAACAGTAAAGATGACATAAAAAGACTGACAGAGGAACTTTGTAAAATAAGTATGAGACTAGAAGAGGTAACTCAATCCTTAAATTACTCTCATCACCGTATGGAAGAACTTATGGCCAAAATTGAGGCCAACGATACCCGTATGAAGTACCTAGAGAAACGGGATCTTGAGTTCACGAATCTTAAATCGGCGGTATCTCAAATACAAAGTGATATAAACATACAGGCTCAAAATATACTGTTAAATGAAATTGAGTTGATTGGAATCCCGGAGTCTCCAGCAGAGAATTTGCAACATACTTTTCAGCTTGCAACTAAGAAGCTGGGTGTCGAACTGGATGAGAAAGATGTAGACTGGATCACTAGGGTGGGGCCTCGCCGTTCTGCAGTTCCTCCTAGCTTAACTGAACTTCAAAATAGGATGCCGCGACCAATTGTTGTTCGTTTGCTGCGTAGATCTAAGCGTGACGAAATACTAAAAGCATCTAAATCACGTAGGAATATAAAAAGTAATGACTTGGATGCCCCTGGAACACCACAAAATATATATTTCAATGAGAGGCTTACTAAAGAAAATAGAATCTTATTTCGAGATGCTAGAAATAGAGCGAAGAAAGAAGGGTACGCTTATTGTTGGTGTAACCGTGGCGCTATATATGTCAGGCGGCGTGAGGGGAATGCAGCAGTGCAACTGCGAAGCCAAAGTTATCTGGATCGGTTATTTGCTAACACGAAAACTACTGTCGGATGTAACAATATCGAGGTATAA
Protein
MKCAGCLKVTKELGSIRCTSSSCTKIFCPLCIDISLVPVDRKKIWKCPDCCAAHKKGGDNSLTPIRGTYDGQNITTRKKEPTAENSKDDIKRLTEELCKISMRLEEVTQSLNYSHHRMEELMAKIEANDTRMKYLEKRDLEFTNLKSAVSQIQSDINIQAQNILLNEIELIGIPESPAENLQHTFQLATKKLGVELDEKDVDWITRVGPRRSAVPPSLTELQNRMPRPIVVRLLRRSKRDEILKASKSRRNIKSNDLDAPGTPQNIYFNERLTKENRILFRDARNRAKKEGYAYCWCNRGAIYVRRREGNAAVQLRSQSYLDRLFANTKTTVGCNNIEV
Summary
Uniprot
A0A0L7L8I5
A0A0L7LA76
A0A0L7KMA8
A0A0L7KR27
A0A2W1BEV5
A0A2H1WRH7
+ More
A0A2A4J6A9 A0A0L7KSF2 A0A2A4J0K9 A0A2A4IZ17 A0A2A4K4X7 A0A2A4J6Z0 A0A2H1WIH0 A0A0L7L550 A0A2W1BHR3 A0A2A4JD92 H9JC60 A0A2H1WVR8 A0A0L7LCM2 A0A2W1C270 A0A2H1WQP2 A0A2A4JDI4 H9JHN2 A0A2A4ITG1 A0A194QNP5 A0A0N0PFG8 A0A0L7KT58 A0A0L7LMB4 A0A0L7LUR8 A0A2A4KAI6 A0A0L7KW99 A0A0L7K2I9 A0A0L7LLW9 A0A2A4J8T1 A0A0L7LT54 A0A0L7LKW3 A0A0L7KNH2 A0A0L7K2H2 A0A3S2TB59 A0A0L7L3H2 A0A0L7LLF8 A0A0L7L9T9 A0A0L7KL91 A0A212F6C5 A0A2A4IUU8 A0A2H1WBH0 A0A2H1X1G7 A0A1Y1JXQ1 A0A1S3DGR2 A0A0L7LJG0 A0A1B6LMZ9 A0A194QEB0 A0A293LWL5 A0A1B6ITU2 A0A2A4JC78 T1HCI2 A0A0L7LF48 T1HA40 A0A0L7K2E3 A0A1S3DD01 A0A0L7L7A2 A0A0L7K4K4 T1HBR1 A0A0L7L864 T1HAF0 A0A0L7K2S8 A0A0A9YWE3 T1HBQ9 A0A0A9YA31 A0A0L7KQ92 A0A194RCQ2 A0A0L7KJT4 A0A2A4IU35 A0A0K8ST40
A0A2A4J6A9 A0A0L7KSF2 A0A2A4J0K9 A0A2A4IZ17 A0A2A4K4X7 A0A2A4J6Z0 A0A2H1WIH0 A0A0L7L550 A0A2W1BHR3 A0A2A4JD92 H9JC60 A0A2H1WVR8 A0A0L7LCM2 A0A2W1C270 A0A2H1WQP2 A0A2A4JDI4 H9JHN2 A0A2A4ITG1 A0A194QNP5 A0A0N0PFG8 A0A0L7KT58 A0A0L7LMB4 A0A0L7LUR8 A0A2A4KAI6 A0A0L7KW99 A0A0L7K2I9 A0A0L7LLW9 A0A2A4J8T1 A0A0L7LT54 A0A0L7LKW3 A0A0L7KNH2 A0A0L7K2H2 A0A3S2TB59 A0A0L7L3H2 A0A0L7LLF8 A0A0L7L9T9 A0A0L7KL91 A0A212F6C5 A0A2A4IUU8 A0A2H1WBH0 A0A2H1X1G7 A0A1Y1JXQ1 A0A1S3DGR2 A0A0L7LJG0 A0A1B6LMZ9 A0A194QEB0 A0A293LWL5 A0A1B6ITU2 A0A2A4JC78 T1HCI2 A0A0L7LF48 T1HA40 A0A0L7K2E3 A0A1S3DD01 A0A0L7L7A2 A0A0L7K4K4 T1HBR1 A0A0L7L864 T1HAF0 A0A0L7K2S8 A0A0A9YWE3 T1HBQ9 A0A0A9YA31 A0A0L7KQ92 A0A194RCQ2 A0A0L7KJT4 A0A2A4IU35 A0A0K8ST40
EMBL
JTDY01002233
KOB71838.1
JTDY01002025
KOB72300.1
JTDY01009015
KOB64201.1
+ More
JTDY01007036 KOB65506.1 KZ150150 PZC72841.1 ODYU01010507 SOQ55661.1 NWSH01002730 PCG67615.1 JTDY01006445 KOB66006.1 NWSH01003975 PCG65605.1 NWSH01005158 PCG64392.1 NWSH01000125 PCG79301.1 NWSH01002711 PCG67639.1 ODYU01008890 SOQ52873.1 JTDY01002966 KOB70404.1 KZ150197 PZC72320.1 NWSH01001982 PCG69514.1 BABH01014938 ODYU01011375 SOQ57082.1 JTDY01001653 KOB73242.1 KZ149908 PZC78213.1 ODYU01010286 SOQ55282.1 NWSH01001817 PCG70031.1 BABH01012931 BABH01012932 NWSH01008646 PCG62462.1 KQ461191 KPJ07137.1 LADJ01030744 KPJ21224.1 JTDY01006205 KOB66199.1 JTDY01000562 KOB76673.1 JTDY01000042 KOB79218.1 NWSH01000010 PCG80924.1 JTDY01005149 KOB67319.1 JTDY01017010 KOB51869.1 JTDY01000624 KOB76430.1 NWSH01002492 PCG68176.1 JTDY01000181 KOB78416.1 JTDY01000729 KOB76082.1 JTDY01007941 KOB64843.1 JTDY01017442 KOB51849.1 RSAL01001410 RVE40802.1 JTDY01003157 KOB70048.1 JTDY01000739 KOB76046.1 JTDY01002139 KOB72021.1 JTDY01009085 KOB64118.1 AGBW02010051 OWR49283.1 NWSH01007173 PCG63054.1 ODYU01007238 SOQ49844.1 ODYU01012716 SOQ59161.1 GEZM01097808 JAV54094.1 JTDY01000945 KOB75331.1 GEBQ01014872 JAT25105.1 KQ459054 KPJ03878.1 GFWV01006510 MAA31240.1 GECU01017357 JAS90349.1 NWSH01002032 PCG69366.1 ACPB03011237 JTDY01001455 KOB73826.1 ACPB03024613 ACPB03027065 JTDY01015045 KOB51976.1 JTDY01006475 JTDY01002579 KOB65982.1 KOB71161.1 JTDY01010989 KOB57787.1 ACPB03014947 JTDY01002313 KOB71687.1 ACPB03021827 JTDY01014864 KOB51988.1 GBHO01009719 GBRD01009686 JAG33885.1 JAG56138.1 ACPB03014935 GBHO01013672 JAG29932.1 JTDY01007401 KOB65231.1 KQ460397 KPJ15249.1 JTDY01009571 KOB63316.1 NWSH01007563 PCG62908.1 GBRD01009465 JAG56359.1
JTDY01007036 KOB65506.1 KZ150150 PZC72841.1 ODYU01010507 SOQ55661.1 NWSH01002730 PCG67615.1 JTDY01006445 KOB66006.1 NWSH01003975 PCG65605.1 NWSH01005158 PCG64392.1 NWSH01000125 PCG79301.1 NWSH01002711 PCG67639.1 ODYU01008890 SOQ52873.1 JTDY01002966 KOB70404.1 KZ150197 PZC72320.1 NWSH01001982 PCG69514.1 BABH01014938 ODYU01011375 SOQ57082.1 JTDY01001653 KOB73242.1 KZ149908 PZC78213.1 ODYU01010286 SOQ55282.1 NWSH01001817 PCG70031.1 BABH01012931 BABH01012932 NWSH01008646 PCG62462.1 KQ461191 KPJ07137.1 LADJ01030744 KPJ21224.1 JTDY01006205 KOB66199.1 JTDY01000562 KOB76673.1 JTDY01000042 KOB79218.1 NWSH01000010 PCG80924.1 JTDY01005149 KOB67319.1 JTDY01017010 KOB51869.1 JTDY01000624 KOB76430.1 NWSH01002492 PCG68176.1 JTDY01000181 KOB78416.1 JTDY01000729 KOB76082.1 JTDY01007941 KOB64843.1 JTDY01017442 KOB51849.1 RSAL01001410 RVE40802.1 JTDY01003157 KOB70048.1 JTDY01000739 KOB76046.1 JTDY01002139 KOB72021.1 JTDY01009085 KOB64118.1 AGBW02010051 OWR49283.1 NWSH01007173 PCG63054.1 ODYU01007238 SOQ49844.1 ODYU01012716 SOQ59161.1 GEZM01097808 JAV54094.1 JTDY01000945 KOB75331.1 GEBQ01014872 JAT25105.1 KQ459054 KPJ03878.1 GFWV01006510 MAA31240.1 GECU01017357 JAS90349.1 NWSH01002032 PCG69366.1 ACPB03011237 JTDY01001455 KOB73826.1 ACPB03024613 ACPB03027065 JTDY01015045 KOB51976.1 JTDY01006475 JTDY01002579 KOB65982.1 KOB71161.1 JTDY01010989 KOB57787.1 ACPB03014947 JTDY01002313 KOB71687.1 ACPB03021827 JTDY01014864 KOB51988.1 GBHO01009719 GBRD01009686 JAG33885.1 JAG56138.1 ACPB03014935 GBHO01013672 JAG29932.1 JTDY01007401 KOB65231.1 KQ460397 KPJ15249.1 JTDY01009571 KOB63316.1 NWSH01007563 PCG62908.1 GBRD01009465 JAG56359.1
Proteomes
Interpro
IPR011011
Znf_FYVE_PHD
+ More
IPR004244 Transposase_22
IPR004941 Baculo_FP
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR002219 PE/DAG-bd
IPR001841 Znf_RING
IPR027267 AH/BAR_dom_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR035979 RBD_domain_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR034209 PUF60_RRM1
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR018527 Rubredoxin_Fe_BS
IPR004244 Transposase_22
IPR004941 Baculo_FP
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR002219 PE/DAG-bd
IPR001841 Znf_RING
IPR027267 AH/BAR_dom_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR035979 RBD_domain_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR034209 PUF60_RRM1
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR018527 Rubredoxin_Fe_BS
SUPFAM
Gene 3D
ProteinModelPortal
A0A0L7L8I5
A0A0L7LA76
A0A0L7KMA8
A0A0L7KR27
A0A2W1BEV5
A0A2H1WRH7
+ More
A0A2A4J6A9 A0A0L7KSF2 A0A2A4J0K9 A0A2A4IZ17 A0A2A4K4X7 A0A2A4J6Z0 A0A2H1WIH0 A0A0L7L550 A0A2W1BHR3 A0A2A4JD92 H9JC60 A0A2H1WVR8 A0A0L7LCM2 A0A2W1C270 A0A2H1WQP2 A0A2A4JDI4 H9JHN2 A0A2A4ITG1 A0A194QNP5 A0A0N0PFG8 A0A0L7KT58 A0A0L7LMB4 A0A0L7LUR8 A0A2A4KAI6 A0A0L7KW99 A0A0L7K2I9 A0A0L7LLW9 A0A2A4J8T1 A0A0L7LT54 A0A0L7LKW3 A0A0L7KNH2 A0A0L7K2H2 A0A3S2TB59 A0A0L7L3H2 A0A0L7LLF8 A0A0L7L9T9 A0A0L7KL91 A0A212F6C5 A0A2A4IUU8 A0A2H1WBH0 A0A2H1X1G7 A0A1Y1JXQ1 A0A1S3DGR2 A0A0L7LJG0 A0A1B6LMZ9 A0A194QEB0 A0A293LWL5 A0A1B6ITU2 A0A2A4JC78 T1HCI2 A0A0L7LF48 T1HA40 A0A0L7K2E3 A0A1S3DD01 A0A0L7L7A2 A0A0L7K4K4 T1HBR1 A0A0L7L864 T1HAF0 A0A0L7K2S8 A0A0A9YWE3 T1HBQ9 A0A0A9YA31 A0A0L7KQ92 A0A194RCQ2 A0A0L7KJT4 A0A2A4IU35 A0A0K8ST40
A0A2A4J6A9 A0A0L7KSF2 A0A2A4J0K9 A0A2A4IZ17 A0A2A4K4X7 A0A2A4J6Z0 A0A2H1WIH0 A0A0L7L550 A0A2W1BHR3 A0A2A4JD92 H9JC60 A0A2H1WVR8 A0A0L7LCM2 A0A2W1C270 A0A2H1WQP2 A0A2A4JDI4 H9JHN2 A0A2A4ITG1 A0A194QNP5 A0A0N0PFG8 A0A0L7KT58 A0A0L7LMB4 A0A0L7LUR8 A0A2A4KAI6 A0A0L7KW99 A0A0L7K2I9 A0A0L7LLW9 A0A2A4J8T1 A0A0L7LT54 A0A0L7LKW3 A0A0L7KNH2 A0A0L7K2H2 A0A3S2TB59 A0A0L7L3H2 A0A0L7LLF8 A0A0L7L9T9 A0A0L7KL91 A0A212F6C5 A0A2A4IUU8 A0A2H1WBH0 A0A2H1X1G7 A0A1Y1JXQ1 A0A1S3DGR2 A0A0L7LJG0 A0A1B6LMZ9 A0A194QEB0 A0A293LWL5 A0A1B6ITU2 A0A2A4JC78 T1HCI2 A0A0L7LF48 T1HA40 A0A0L7K2E3 A0A1S3DD01 A0A0L7L7A2 A0A0L7K4K4 T1HBR1 A0A0L7L864 T1HAF0 A0A0L7K2S8 A0A0A9YWE3 T1HBQ9 A0A0A9YA31 A0A0L7KQ92 A0A194RCQ2 A0A0L7KJT4 A0A2A4IU35 A0A0K8ST40
Ontologies
KEGG
PANTHER
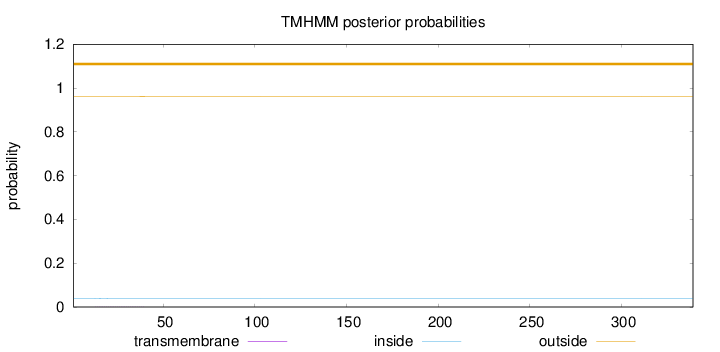
Topology
Length:
339
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02321
Exp number, first 60 AAs:
0.02321
Total prob of N-in:
0.03995
outside
1 - 339
Population Genetic Test Statistics
Pi
13.649484
Theta
11.775169
Tajima's D
-0.685126
CLR
17.706871
CSRT
0.19894005299735
Interpretation
Uncertain
