Pre Gene Modal
BGIBMGA012059
Annotation
PREDICTED:_U3_small_nucleolar_RNA-associated_protein_6_homolog_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.911
Sequence
CDS
ATGGCTGAACAAGTAAATCAACGTATTGAGAATATGATAAATGAGTTGGAGCAAATGAGAAGAACAAACTTATATACAGATGAAGAGATAAAAGAAATATCCCAAAAACGAAAAGAGTTTGAATATCACATACAAAGGAGAGTCAAACAAAAAGAAGATTATGTTCAATACATCGCTTATGAACTTGCCTTACTTGAGGATATAACAACGCGTAGAAAAAAAATACAACTAACAGAGAAGAAAAAGGATTTGGAGTATGCCATTGCTCAACGTTTGAACCAACTTTTCAAGCGCTTTATTTATAAGTTCCAAAATGAAATTGAAATATATTTTGAGTATATAAAATTCTGTAAGGGTGTGGGGTTTCAACAGGCTATCTCAGGTATCATCGGACAAATGTTGCAGATACATGGTGATAAACCTCAAATGTGGTTGATGGCGAGCAAATGGGAGAGTTTAGAACAGAACAATCTAGAAAATGCAAAGGCCTTTCTTTTAAAGGGAATTCAAAGAAACCCTGATGCTGAGCCATTGTACCTTGAATTGTTCAACATTGAGCTAATTGATTTGTGCTTTAAGGCGGAGACTGACGAAGAAAAGGAGAAACTGCAAAAGAAGGCTGATATAATCTGGAGAAACGGAGCAAAGAATATCCAAAATGTGAAGTTTCTATTCAAATTGTATGACTTGTGTACCAAGTATGAATATACCGGTGGATTAGTGGACGATATAAAACTTGAAATATGGTCAAATCGTAATAACCAAGAAGTTTGGCCTTATCTTGCATTGAAGGAACTGGAAGGTTTTCATTGGAAGGAAATAGAGGAATTTGCTGATGATGAACATAATTTTCCTAAAGTTGTTGGTTATGTAATAGGTATTTACGAAGAAGCACTGCAATTGTTTATGGACGAAAATTTATGCACACGTTTCATCCATGATTTGCTTGGTATGAGTGAAGATATATGTTCCGATCAACAAAAAATCAAAGCTGTAAAACATGCTTGGATGTACGGTCATGACAATGGTCTTCTCAGCAATGACATGTATGCGTTTGGCATTGAAATGTTGAAACTGGAGAATGAGACAACTACTGATGAACTATTAGAGATCTTAAATGCAGCAATAAAGAGAAATCCCAAACTAAAGAGTGTCTGGGAACAGTTGTTACTTCATAAAAAAGACGATGAGAAACAACTTTTAGTAATACTACAAAATGCAACAAAGTCTTTGAATACTGGTGATGCCTTGCAGTTGTATAATATTCTATTAGATAACATTGAATCAGGGCCTATGATCAAAACCCTATACAAGAAGTTTCAAAATTGTGACAATGCAATTCTTCTTGCTGTTAAACCAAAACTTTTACAGAAAATGTACGAACATAATGGCCTGAAAGCTACTAGGGAATTGTATGAAGATTTTATTAGGACACCACCAACACAAATTGAACTTCATAAAATAATGATCAATATCGAAAAATCGCAAGAAAAGCTAAATTTGAAAGCAATTAGAAAATGTTATGAGTGTGCTGTGCATCATCACGGGACTGACAACAAAGAAATTTGGGCAGATTATATTAATTTTGAAATTGAGAATGGGAATGCTCCAAATTCAGCAGTTGTATATAGAAGAGCTGTGGGCATGCTCAAGAAAGATATTGTTGATGAGTTTATTAGAGATCAAACTTTATCAAAAATAAAATGA
Protein
MAEQVNQRIENMINELEQMRRTNLYTDEEIKEISQKRKEFEYHIQRRVKQKEDYVQYIAYELALLEDITTRRKKIQLTEKKKDLEYAIAQRLNQLFKRFIYKFQNEIEIYFEYIKFCKGVGFQQAISGIIGQMLQIHGDKPQMWLMASKWESLEQNNLENAKAFLLKGIQRNPDAEPLYLELFNIELIDLCFKAETDEEKEKLQKKADIIWRNGAKNIQNVKFLFKLYDLCTKYEYTGGLVDDIKLEIWSNRNNQEVWPYLALKELEGFHWKEIEEFADDEHNFPKVVGYVIGIYEEALQLFMDENLCTRFIHDLLGMSEDICSDQQKIKAVKHAWMYGHDNGLLSNDMYAFGIEMLKLENETTTDELLEILNAAIKRNPKLKSVWEQLLLHKKDDEKQLLVILQNATKSLNTGDALQLYNILLDNIESGPMIKTLYKKFQNCDNAILLAVKPKLLQKMYEHNGLKATRELYEDFIRTPPTQIELHKIMINIEKSQEKLNLKAIRKCYECAVHHHGTDNKEIWADYINFEIENGNAPNSAVVYRRAVGMLKKDIVDEFIRDQTLSKIK
Summary
Uniprot
A0A2A4JU10
A0A2H1W965
A0A1E1WCU7
A0A194PFF5
A0A194R487
A0A3S2NWH1
+ More
A0A212ERY2 A0A0L7LAS7 A0A1E1WND8 A0A067RGS1 A0A0L7QXW4 A0A2J7QS59 E2BZ17 A0A0C9QS71 A0A087ZQI1 A0A2A3E472 A0A232ETI9 K7JA79 A0A195CXK4 E2AAN7 T1HPQ3 A0A154P8Y7 E2BZ18 A0A0P4VPP8 A0A224XA13 A0A195FX33 A0A195ATW6 A0A023FA15 A0A0V0G6L9 F4WUU5 A0A151JCL0 A0A158P0T0 A0A336LW32 A0A151WW60 A0A3L8E0X1 H9GF89 A0A1L8DFJ2 R4WST9 E9IZA8 U3FZ92 A0A2R5L4Z8 A0A0F7Z2C8 A0A026VWK0 A0A023GLK8 U3JHX0 H0Z3L4 A0A1B0CGG0 A0A1B6E402 A0A1B6HLF9 A0A1W7RBR5 A0A1B6GID5 A0A218UWU7 F1NLG7 A0A1S3IB83 G3MNC1 A0A3L8SBT8 A0A293MWJ1 A0A2D4H9X5 A0A147BSV4 A0A1U8DSY5 A0A0Q3TFY5 A0A1U7RND9 A0A131XVT8 A0A091F0S4 E9GVM3 W5UE51 E0W1D1 A0A1E1XR21 A0A1A7WJX9 A0A099ZDL5 A0A3P8XPJ1 A0A1E1X2J2 A0A1V4KW80 A0A131XLP9 A0A1Z5KVB9 H3BC90 A0A224Z2S1 A0A060XD50 A0A131YT66 A0A0C9PQJ7 A0A3B4EJ79 A0A3Q2GPQ8 K7G189 A0A2R8RMC9 A0A3Q1BIA4 A0A0R4IGM5 A9JTC5 A0A1A8GSY8 A0A1U7QXW4 A0A1A8AL68 A0A3P8U3X4 L7M6U4 A0A1A8LL24 A0A3Q2W658 A0A3P8NYI3 A0A3P9DBZ2 Q803K2
A0A212ERY2 A0A0L7LAS7 A0A1E1WND8 A0A067RGS1 A0A0L7QXW4 A0A2J7QS59 E2BZ17 A0A0C9QS71 A0A087ZQI1 A0A2A3E472 A0A232ETI9 K7JA79 A0A195CXK4 E2AAN7 T1HPQ3 A0A154P8Y7 E2BZ18 A0A0P4VPP8 A0A224XA13 A0A195FX33 A0A195ATW6 A0A023FA15 A0A0V0G6L9 F4WUU5 A0A151JCL0 A0A158P0T0 A0A336LW32 A0A151WW60 A0A3L8E0X1 H9GF89 A0A1L8DFJ2 R4WST9 E9IZA8 U3FZ92 A0A2R5L4Z8 A0A0F7Z2C8 A0A026VWK0 A0A023GLK8 U3JHX0 H0Z3L4 A0A1B0CGG0 A0A1B6E402 A0A1B6HLF9 A0A1W7RBR5 A0A1B6GID5 A0A218UWU7 F1NLG7 A0A1S3IB83 G3MNC1 A0A3L8SBT8 A0A293MWJ1 A0A2D4H9X5 A0A147BSV4 A0A1U8DSY5 A0A0Q3TFY5 A0A1U7RND9 A0A131XVT8 A0A091F0S4 E9GVM3 W5UE51 E0W1D1 A0A1E1XR21 A0A1A7WJX9 A0A099ZDL5 A0A3P8XPJ1 A0A1E1X2J2 A0A1V4KW80 A0A131XLP9 A0A1Z5KVB9 H3BC90 A0A224Z2S1 A0A060XD50 A0A131YT66 A0A0C9PQJ7 A0A3B4EJ79 A0A3Q2GPQ8 K7G189 A0A2R8RMC9 A0A3Q1BIA4 A0A0R4IGM5 A9JTC5 A0A1A8GSY8 A0A1U7QXW4 A0A1A8AL68 A0A3P8U3X4 L7M6U4 A0A1A8LL24 A0A3Q2W658 A0A3P8NYI3 A0A3P9DBZ2 Q803K2
Pubmed
26354079
22118469
26227816
24845553
20798317
28648823
+ More
20075255 27129103 25474469 21719571 21347285 30249741 21881562 23691247 21282665 23915248 24508170 20360741 26358130 15592404 22216098 30282656 29652888 21292972 23127152 20566863 29209593 25069045 28503490 28049606 28528879 9215903 28797301 24755649 26830274 17381049 23594743 25576852 25186727
20075255 27129103 25474469 21719571 21347285 30249741 21881562 23691247 21282665 23915248 24508170 20360741 26358130 15592404 22216098 30282656 29652888 21292972 23127152 20566863 29209593 25069045 28503490 28049606 28528879 9215903 28797301 24755649 26830274 17381049 23594743 25576852 25186727
EMBL
NWSH01000618
PCG75266.1
ODYU01007134
SOQ49639.1
GDQN01006246
JAT84808.1
+ More
KQ459606 KPI91753.1 KQ460779 KPJ12334.1 RSAL01000045 RVE50627.1 AGBW02012898 OWR44253.1 JTDY01002015 KOB72321.1 GDQN01002540 JAT88514.1 KK852649 KDR19476.1 KQ414699 KOC63457.1 NEVH01011878 PNF31407.1 GL451549 EFN79056.1 GBYB01003497 JAG73264.1 KZ288386 PBC26488.1 NNAY01002265 OXU21679.1 AAZX01008028 KQ977141 KYN05376.1 GL438137 EFN69487.1 ACPB03001895 KQ434846 KZC08369.1 EFN79057.1 GDKW01001618 JAI54977.1 GFTR01007229 JAW09197.1 KQ981200 KYN44998.1 KQ976738 KYM75673.1 GBBI01000390 JAC18322.1 GECL01002385 JAP03739.1 GL888378 EGI62009.1 KQ979038 KYN23468.1 ADTU01005780 ADTU01005781 UFQS01000144 UFQT01000144 SSX00473.1 SSX20853.1 KQ982691 KYQ52068.1 QOIP01000001 RLU26381.1 AAWZ02011871 GFDF01008852 JAV05232.1 AK417782 BAN20997.1 GL767121 EFZ14077.1 GAEP01001041 JAB53780.1 GGLE01000391 MBY04517.1 GBEX01003779 JAI10781.1 KK107921 EZA47219.1 GBBM01000689 JAC34729.1 AGTO01011150 ABQF01037331 AJWK01011081 GEDC01021530 GEDC01004632 JAS15768.1 JAS32666.1 GECU01032187 GECU01010668 JAS75519.1 JAS97038.1 GDAY02002883 JAV48584.1 GECZ01007584 JAS62185.1 MUZQ01000104 OWK58275.1 AADN05000628 JO843372 AEO34989.1 QUSF01000033 RLV99258.1 GFWV01018972 MAA43700.1 IACK01020264 LAA68780.1 GEGO01001949 JAR93455.1 LMAW01002589 KQK79454.1 GEFM01006010 JAP69786.1 KK719227 KFO62129.1 GL732568 EFX76504.1 JT413274 AHH40180.1 DS235869 EEB19437.1 GFAA01001672 JAU01763.1 HADW01004439 HADX01010770 SBP05839.1 KL892480 KGL79867.1 GFAC01005703 JAT93485.1 LSYS01001520 OPJ88561.1 GEFH01002130 JAP66451.1 GFJQ02007918 JAV99051.1 AFYH01051099 AFYH01051100 GFPF01013012 MAA24158.1 FR904998 CDQ74760.1 GEDV01006877 JAP81680.1 GBYB01003498 JAG73265.1 AGCU01100770 AGCU01100771 AGCU01100772 BX663498 AL954746 CR589950 BC155285 AAI55286.1 HAEB01013598 HAEC01006205 SBQ74282.1 HADY01016475 HAEJ01015245 SBP54960.1 GACK01005018 JAA60016.1 HAEF01007207 HAEG01013045 SBR44589.1 BC044444 AAH44444.1
KQ459606 KPI91753.1 KQ460779 KPJ12334.1 RSAL01000045 RVE50627.1 AGBW02012898 OWR44253.1 JTDY01002015 KOB72321.1 GDQN01002540 JAT88514.1 KK852649 KDR19476.1 KQ414699 KOC63457.1 NEVH01011878 PNF31407.1 GL451549 EFN79056.1 GBYB01003497 JAG73264.1 KZ288386 PBC26488.1 NNAY01002265 OXU21679.1 AAZX01008028 KQ977141 KYN05376.1 GL438137 EFN69487.1 ACPB03001895 KQ434846 KZC08369.1 EFN79057.1 GDKW01001618 JAI54977.1 GFTR01007229 JAW09197.1 KQ981200 KYN44998.1 KQ976738 KYM75673.1 GBBI01000390 JAC18322.1 GECL01002385 JAP03739.1 GL888378 EGI62009.1 KQ979038 KYN23468.1 ADTU01005780 ADTU01005781 UFQS01000144 UFQT01000144 SSX00473.1 SSX20853.1 KQ982691 KYQ52068.1 QOIP01000001 RLU26381.1 AAWZ02011871 GFDF01008852 JAV05232.1 AK417782 BAN20997.1 GL767121 EFZ14077.1 GAEP01001041 JAB53780.1 GGLE01000391 MBY04517.1 GBEX01003779 JAI10781.1 KK107921 EZA47219.1 GBBM01000689 JAC34729.1 AGTO01011150 ABQF01037331 AJWK01011081 GEDC01021530 GEDC01004632 JAS15768.1 JAS32666.1 GECU01032187 GECU01010668 JAS75519.1 JAS97038.1 GDAY02002883 JAV48584.1 GECZ01007584 JAS62185.1 MUZQ01000104 OWK58275.1 AADN05000628 JO843372 AEO34989.1 QUSF01000033 RLV99258.1 GFWV01018972 MAA43700.1 IACK01020264 LAA68780.1 GEGO01001949 JAR93455.1 LMAW01002589 KQK79454.1 GEFM01006010 JAP69786.1 KK719227 KFO62129.1 GL732568 EFX76504.1 JT413274 AHH40180.1 DS235869 EEB19437.1 GFAA01001672 JAU01763.1 HADW01004439 HADX01010770 SBP05839.1 KL892480 KGL79867.1 GFAC01005703 JAT93485.1 LSYS01001520 OPJ88561.1 GEFH01002130 JAP66451.1 GFJQ02007918 JAV99051.1 AFYH01051099 AFYH01051100 GFPF01013012 MAA24158.1 FR904998 CDQ74760.1 GEDV01006877 JAP81680.1 GBYB01003498 JAG73265.1 AGCU01100770 AGCU01100771 AGCU01100772 BX663498 AL954746 CR589950 BC155285 AAI55286.1 HAEB01013598 HAEC01006205 SBQ74282.1 HADY01016475 HAEJ01015245 SBP54960.1 GACK01005018 JAA60016.1 HAEF01007207 HAEG01013045 SBR44589.1 BC044444 AAH44444.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
UP000037510
+ More
UP000027135 UP000053825 UP000235965 UP000008237 UP000005203 UP000242457 UP000215335 UP000002358 UP000078542 UP000000311 UP000015103 UP000076502 UP000078541 UP000078540 UP000007755 UP000078492 UP000005205 UP000075809 UP000279307 UP000001646 UP000053097 UP000016665 UP000007754 UP000092461 UP000197619 UP000000539 UP000085678 UP000276834 UP000189705 UP000051836 UP000052976 UP000000305 UP000221080 UP000009046 UP000053641 UP000265140 UP000190648 UP000008672 UP000193380 UP000261440 UP000265020 UP000007267 UP000000437 UP000257160 UP000189706 UP000265080 UP000264840 UP000265100 UP000265160
UP000027135 UP000053825 UP000235965 UP000008237 UP000005203 UP000242457 UP000215335 UP000002358 UP000078542 UP000000311 UP000015103 UP000076502 UP000078541 UP000078540 UP000007755 UP000078492 UP000005205 UP000075809 UP000279307 UP000001646 UP000053097 UP000016665 UP000007754 UP000092461 UP000197619 UP000000539 UP000085678 UP000276834 UP000189705 UP000051836 UP000052976 UP000000305 UP000221080 UP000009046 UP000053641 UP000265140 UP000190648 UP000008672 UP000193380 UP000261440 UP000265020 UP000007267 UP000000437 UP000257160 UP000189706 UP000265080 UP000264840 UP000265100 UP000265160
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A2A4JU10
A0A2H1W965
A0A1E1WCU7
A0A194PFF5
A0A194R487
A0A3S2NWH1
+ More
A0A212ERY2 A0A0L7LAS7 A0A1E1WND8 A0A067RGS1 A0A0L7QXW4 A0A2J7QS59 E2BZ17 A0A0C9QS71 A0A087ZQI1 A0A2A3E472 A0A232ETI9 K7JA79 A0A195CXK4 E2AAN7 T1HPQ3 A0A154P8Y7 E2BZ18 A0A0P4VPP8 A0A224XA13 A0A195FX33 A0A195ATW6 A0A023FA15 A0A0V0G6L9 F4WUU5 A0A151JCL0 A0A158P0T0 A0A336LW32 A0A151WW60 A0A3L8E0X1 H9GF89 A0A1L8DFJ2 R4WST9 E9IZA8 U3FZ92 A0A2R5L4Z8 A0A0F7Z2C8 A0A026VWK0 A0A023GLK8 U3JHX0 H0Z3L4 A0A1B0CGG0 A0A1B6E402 A0A1B6HLF9 A0A1W7RBR5 A0A1B6GID5 A0A218UWU7 F1NLG7 A0A1S3IB83 G3MNC1 A0A3L8SBT8 A0A293MWJ1 A0A2D4H9X5 A0A147BSV4 A0A1U8DSY5 A0A0Q3TFY5 A0A1U7RND9 A0A131XVT8 A0A091F0S4 E9GVM3 W5UE51 E0W1D1 A0A1E1XR21 A0A1A7WJX9 A0A099ZDL5 A0A3P8XPJ1 A0A1E1X2J2 A0A1V4KW80 A0A131XLP9 A0A1Z5KVB9 H3BC90 A0A224Z2S1 A0A060XD50 A0A131YT66 A0A0C9PQJ7 A0A3B4EJ79 A0A3Q2GPQ8 K7G189 A0A2R8RMC9 A0A3Q1BIA4 A0A0R4IGM5 A9JTC5 A0A1A8GSY8 A0A1U7QXW4 A0A1A8AL68 A0A3P8U3X4 L7M6U4 A0A1A8LL24 A0A3Q2W658 A0A3P8NYI3 A0A3P9DBZ2 Q803K2
A0A212ERY2 A0A0L7LAS7 A0A1E1WND8 A0A067RGS1 A0A0L7QXW4 A0A2J7QS59 E2BZ17 A0A0C9QS71 A0A087ZQI1 A0A2A3E472 A0A232ETI9 K7JA79 A0A195CXK4 E2AAN7 T1HPQ3 A0A154P8Y7 E2BZ18 A0A0P4VPP8 A0A224XA13 A0A195FX33 A0A195ATW6 A0A023FA15 A0A0V0G6L9 F4WUU5 A0A151JCL0 A0A158P0T0 A0A336LW32 A0A151WW60 A0A3L8E0X1 H9GF89 A0A1L8DFJ2 R4WST9 E9IZA8 U3FZ92 A0A2R5L4Z8 A0A0F7Z2C8 A0A026VWK0 A0A023GLK8 U3JHX0 H0Z3L4 A0A1B0CGG0 A0A1B6E402 A0A1B6HLF9 A0A1W7RBR5 A0A1B6GID5 A0A218UWU7 F1NLG7 A0A1S3IB83 G3MNC1 A0A3L8SBT8 A0A293MWJ1 A0A2D4H9X5 A0A147BSV4 A0A1U8DSY5 A0A0Q3TFY5 A0A1U7RND9 A0A131XVT8 A0A091F0S4 E9GVM3 W5UE51 E0W1D1 A0A1E1XR21 A0A1A7WJX9 A0A099ZDL5 A0A3P8XPJ1 A0A1E1X2J2 A0A1V4KW80 A0A131XLP9 A0A1Z5KVB9 H3BC90 A0A224Z2S1 A0A060XD50 A0A131YT66 A0A0C9PQJ7 A0A3B4EJ79 A0A3Q2GPQ8 K7G189 A0A2R8RMC9 A0A3Q1BIA4 A0A0R4IGM5 A9JTC5 A0A1A8GSY8 A0A1U7QXW4 A0A1A8AL68 A0A3P8U3X4 L7M6U4 A0A1A8LL24 A0A3Q2W658 A0A3P8NYI3 A0A3P9DBZ2 Q803K2
PDB
5WLC
E-value=7.16749e-15,
Score=198
Ontologies
GO
Topology
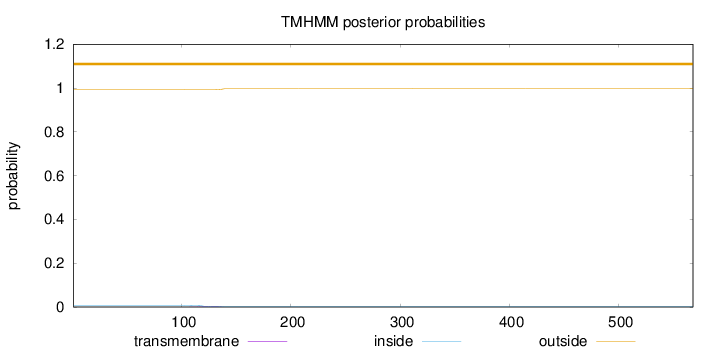
Length:
568
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0890299999999999
Exp number, first 60 AAs:
0.00057
Total prob of N-in:
0.00810
outside
1 - 568
Population Genetic Test Statistics
Pi
32.10366
Theta
29.119574
Tajima's D
0.280458
CLR
0.699429
CSRT
0.461026948652567
Interpretation
Uncertain
