Gene
KWMTBOMO06834
Pre Gene Modal
BGIBMGA012058
Annotation
PREDICTED:_tonsoku-like_protein_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.26
Sequence
CDS
ATGGAAGAAGAAAAACTTTTGCGTAAAAAAAAGAAAGCTCTAAACGGAAGTAACAGAAGAGCCTTAGCAGAAGCTTGTAATGATTTAGCTACGTTTTATTACAAAAATAATCGGTACAATGACGCTCTCGACGAGTATAAAATTGAAGCCTCGGTATGCAAAGAGTTGGGTCTCCGTATGGAATGGGGAACTTGCAGCAGGATGATCGGCGAGATGTACATGTTAATGGCTGAATTTGATAAAGCCCTGAAATACGAAGAGAGACATTTGGCTGTAGCTAAAGAATTAAAGAATTTAGTGGAAGAACAAAGAGCTATGGCAACATTAGGCAGGATATATCTGCTACAAGGACAAAGCACAATTGATGAAGTTGAGGCAAAGACATCTCTGAATGCAGCAGAAAAAGCTTTCATGAAAAGCTTAGTGTTGTGTGAAAATCTAAATGGAAAAATAAATAAACATGAGTTAATGGACATGAGAGCCCGATTACTCCTGAATATTGGTGTGGTCCAAGAACACCTTGGTTTTCTTGAAAAAGCAACAGATTGTATACATAAAGCAATCTCAATATGTTCTAATCAGGACTTGTATGAAGTATTGCATAATTGTTATATGACTGAAGCATTGCTACATAGCAATAAAAAGAAGGATTATGCAAAAGCTTTAAGTTGCTTGAATAAAGCCCTTGAAATAGCAAGTAGACTGGAGAATAAGGTTCTGAAGACGTGTGAAACACTATCATCAAAAGCAGACATACTGTGCAAAATCTCAGACTATCAGAGTGCCAAACAAGTGCTCCTCAAAGCATGGAAGTTGAAGACCCCTGATGAAGAAGAAAGGGATAATATTGAAACAAATTTGAGAGTTGTGGCGGCAATGTGCTATACAGAAGATCTCTTAATCTCAACGAACCCATCAGACCACGCGACCTTCAAGAAACTTTATGAGAAGCTTGGAGATGGAGCTTGTCACATTCGGAATTATCCCGGGGCCGTAGAGTATTATCTGAAGATGTTGGAACACGCTGAACTAGCTGGAGATTGTGGAAAGACTTTAATACCCATTTATGTTAGCTTGTATCAAACATACAAAGACATGGGTTTGTATAGTGAAGCCTTAGATTATTACGAGAAAGAATACGAACTTATAAAGGATGTCCCGAAAGAAGCATACACTACACTCTACAACATTGCGGAGACATTGTATCTGGCTAAGAGGCCTTATGAACAAGTAGAAGAGGCATGTTTAAATGCTCGGAAAGCTGCACAAGCTTGGAATAAAAGGAAATATGAAATCAGAATATTAAGAAATCTTCTAAAATACCAAGAGGCATATTTTGAGATTGATAAAACGGAAGAGACCAAATATGAACTAAGAACATTAGGCTGTGATAATTTTGACAACGTGGAGCATTCCGAAGATGATCAAAGTTCAGCGGGTGGGGGGGACGATGACACGACTCATATCGGTGATGATATATGCCTCGATGATCTTACAGATATGTCTGACACAAATGAAGAAGACGTGACAGAGACGAAACGCGAGACAAGAAAGAGAGGGAAGGGCTATGTTATAAAGAAAAATATGAAAGGCGAAACACAGTTACATGTAGCTGCTATATCCGGAAACAAACTTCTGTTGGAACGGCTTCTAGCCAAAGGACATCCGGTCAATATCAGGGACAATGCCGGCTGGCTACCCCTACACGAGGCTTGCATTCATGGGCACCTTGAAATAGCTAACATGCTCATACACAACGGAGCCAATGTCAATGATCGGGGCGGCTCCAACTGTGATGGTATTACCCCGCTGTATGACGCCGCCAGTAATGGTCATCTCGAAGTCGTACAGCTACTCCTAGAAAAGGGCGCGATACCTTCACTGAAGACAGACTTCGGGGACACGCCCTTAAGGGCACTGCAGAAATGGCGGGCCGGCACTATCCTCACCAGAGACGAAGAAACCATGTACAATAACGTGTGCAATAAAATCCAAAATTCAAGCGATAGAACCAACGCTGACCTAAGGTGCAAATCGGTGACTCCGGTCAAAAATGAAGACGAAATCGCTCCGCCAAGCACGTCAAAATCCAAGAGGAATCTAGACTCACCGGCGCTCAGACGCAGAAATATCATTGATGACGGCAGCGATGACGAAATCGATCTGTCTCAGAACGTCCGGAGAGAATTGGCGTTCCCGTCCGACGATTCAAACGATTCCGACGAAAAATCAAACGTTAATATTGAGGTTTCCGGTGTCAAAGAGTACAGGAATGCCATGTCCGCTTTGAGAAACAGAAACCTCAACGACCATTCTGAAGTTAATACGAAAAAGACGAAGCCTAAGTCAGCTCTGTTAGCCCCCGAAGAGGTCGACGATGATTGGTTGGAAGACGATTTGGGTATTCATAAAAATAAAAAGAGAAAACTCAGCGACCCACTCACGATCGTCTCGAAGAAACCTTCGTACGAACAAATCAAGGAAAGCGTTGACAGTATCAATAGCATACCAGAACCTCTGATCGAAAATACCTTGAAACAAAACAGGTCACGGATCGATGACGTCATCGATATAGACTATATCTCGGATTACGATCGGGTAGACGAGAACATTCGACCCGAAGCAAGTCCCGCCAAATTCGCGAGGAACAATACAAGAGAATTAGATTTTAAGACGAACGACAGCAGGGAGAACATGAGGAAACGCTGGAAGCGCCAGAGCACGCTACTGAAGTCCGGCTTCCAGAGGAAACTGGACGAGGACCAATCTAGTAACAGCGGAAGTGAAACCGAATTTACTAGAAGAGACTCTTTTCATGCTAGAAACAAGTCGATCTCCCGACATTCTTCCGCCGAAAATTTCAATGAAGGCTTCAATATAATGCAAAATATTAATCCTAATATCGTCCAACCGGTAAATGTGATACAGCCAATTAATATAGTCAAATCGACGAAGAACGGGAGGCCGGTACAGACGCAGATACTACCACCGGCCGCCGTGAAGGTTCACATAGAGGACAAAGTGTTGCTGATCTCTTTGAAATTGGACACAATAAACAGATTGACGATAAGCTGGCTGGTTGAGGAAGTCAAGACCAGATATTATAAACTGACCGGCGTGCGTCCAGTGTTCAGCCTGATGACGTCAGACGGTGCGATTCTCTCCGAAGACGACCCCCTGAGCCTGGTGCTCTCGTCGCCCGAACTGAAGACCTGCATCACCAACTGGAAGGCTTCTCCAGCTGAAGAACGATACTTGGAGTGCTGTGATGCTTTGGGAACGTCGCCCTCGAAGGAAATTCAAGAGGCAATCGGCCGGAGTCTCACGACTCGCAGACTGGCGCTCGGTCCGCGGACGCTCTCCTCCTCGCAAACACGACCCCTCTTCAGAGCGATCACCCACCAAACACACATCACGGCCATCATAATGTCGGACAACAATTTGGGAGACGCCGGCGTCAAACATCTAGTTGATTGTGTGTCGACATTGAAACACCTGACGCTGATTGATATCAGCAAAAACAATATCTCCAGTGAAGGAACGAAATTGATACTGAACTTGTTCGAGAAATCGCCCAGAATGGTGTGTCAATCGTTAGAGGAATTTGTAATTAGCAACAATCCAATTTCTGATGACGGATTCCGGAATATAATCAGAATGTTTGATTTGGAGTTGAACAATGTGGGATTGGAAGGGAGTGCCGTTGGATGTGTTGCCAGCTTTCTGGATTCTGCTAAAGATTTAAAGTTGAGGCGCTTTGGTTTATCGAACTGTAAACTCGTCGATGGACAGTTTATGAGGATATTCAGAAGCCTGAGCAGAGCCAAACACCTACAATCAATAAGTCTAAGGAAAAACAACATAACGTTTATAACACTCAAGAAACTGCTACAAAGACAGCCGCCCGTGCCACAAGTCAACTTAGAAGGGTGCCCGGACATATTCAAGTACTCGCCCGATTCAGATTTTCAAGTGTGGCTACCAGCAGTCAACTTTGGACGGTGTTTACCAGAAATCAATGTCACCCCAGTGTCTAAGAGCGAGGAGGAGAGAGACAGTTTTAAGTCTTTCTCTAAAACCTGGCTCAGTTGTTTCAATGGTAGGGGTGTCATAGAACATTGCGATGGAACAGTAATCAAATTAACGGCTAGGTGA
Protein
MEEEKLLRKKKKALNGSNRRALAEACNDLATFYYKNNRYNDALDEYKIEASVCKELGLRMEWGTCSRMIGEMYMLMAEFDKALKYEERHLAVAKELKNLVEEQRAMATLGRIYLLQGQSTIDEVEAKTSLNAAEKAFMKSLVLCENLNGKINKHELMDMRARLLLNIGVVQEHLGFLEKATDCIHKAISICSNQDLYEVLHNCYMTEALLHSNKKKDYAKALSCLNKALEIASRLENKVLKTCETLSSKADILCKISDYQSAKQVLLKAWKLKTPDEEERDNIETNLRVVAAMCYTEDLLISTNPSDHATFKKLYEKLGDGACHIRNYPGAVEYYLKMLEHAELAGDCGKTLIPIYVSLYQTYKDMGLYSEALDYYEKEYELIKDVPKEAYTTLYNIAETLYLAKRPYEQVEEACLNARKAAQAWNKRKYEIRILRNLLKYQEAYFEIDKTEETKYELRTLGCDNFDNVEHSEDDQSSAGGGDDDTTHIGDDICLDDLTDMSDTNEEDVTETKRETRKRGKGYVIKKNMKGETQLHVAAISGNKLLLERLLAKGHPVNIRDNAGWLPLHEACIHGHLEIANMLIHNGANVNDRGGSNCDGITPLYDAASNGHLEVVQLLLEKGAIPSLKTDFGDTPLRALQKWRAGTILTRDEETMYNNVCNKIQNSSDRTNADLRCKSVTPVKNEDEIAPPSTSKSKRNLDSPALRRRNIIDDGSDDEIDLSQNVRRELAFPSDDSNDSDEKSNVNIEVSGVKEYRNAMSALRNRNLNDHSEVNTKKTKPKSALLAPEEVDDDWLEDDLGIHKNKKRKLSDPLTIVSKKPSYEQIKESVDSINSIPEPLIENTLKQNRSRIDDVIDIDYISDYDRVDENIRPEASPAKFARNNTRELDFKTNDSRENMRKRWKRQSTLLKSGFQRKLDEDQSSNSGSETEFTRRDSFHARNKSISRHSSAENFNEGFNIMQNINPNIVQPVNVIQPINIVKSTKNGRPVQTQILPPAAVKVHIEDKVLLISLKLDTINRLTISWLVEEVKTRYYKLTGVRPVFSLMTSDGAILSEDDPLSLVLSSPELKTCITNWKASPAEERYLECCDALGTSPSKEIQEAIGRSLTTRRLALGPRTLSSSQTRPLFRAITHQTHITAIIMSDNNLGDAGVKHLVDCVSTLKHLTLIDISKNNISSEGTKLILNLFEKSPRMVCQSLEEFVISNNPISDDGFRNIIRMFDLELNNVGLEGSAVGCVASFLDSAKDLKLRRFGLSNCKLVDGQFMRIFRSLSRAKHLQSISLRKNNITFITLKKLLQRQPPVPQVNLEGCPDIFKYSPDSDFQVWLPAVNFGRCLPEINVTPVSKSEEERDSFKSFSKTWLSCFNGRGVIEHCDGTVIKLTAR
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Uniprot
EMBL
BABH01016216
RSAL01000045
RVE50626.1
KQ459606
KPI91757.1
KQ460779
+ More
KPJ12339.1 AGBW02012898 OWR44254.1 ODYU01007134 SOQ49640.1 JTDY01008094 KOB64744.1 CM002912 KMY98198.1 KMY98199.1 CH379069 EAL30762.1 CH479188 EDW40122.1 CM000159 EDW93794.1 CH964168 EDW80104.1 CH940647 EDW70531.2 UFQS01000219 UFQT01000219 SSX01535.1 SSX21915.1 LJIG01016206 KRT81298.1 KRF84961.1
KPJ12339.1 AGBW02012898 OWR44254.1 ODYU01007134 SOQ49640.1 JTDY01008094 KOB64744.1 CM002912 KMY98198.1 KMY98199.1 CH379069 EAL30762.1 CH479188 EDW40122.1 CM000159 EDW93794.1 CH964168 EDW80104.1 CH940647 EDW70531.2 UFQS01000219 UFQT01000219 SSX01535.1 SSX21915.1 LJIG01016206 KRT81298.1 KRF84961.1
Proteomes
PRIDE
Interpro
IPR019734
TPR_repeat
+ More
IPR001611 Leu-rich_rpt
IPR011990 TPR-like_helical_dom_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR032675 LRR_dom_sf
IPR013026 TPR-contain_dom
IPR006553 Leu-rich_rpt_Cys-con_subtyp
IPR001995 Peptidase_A2_cat
IPR008952 Tetraspanin_EC2_sf
IPR018499 Tetraspanin/Peripherin
IPR001611 Leu-rich_rpt
IPR011990 TPR-like_helical_dom_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR032675 LRR_dom_sf
IPR013026 TPR-contain_dom
IPR006553 Leu-rich_rpt_Cys-con_subtyp
IPR001995 Peptidase_A2_cat
IPR008952 Tetraspanin_EC2_sf
IPR018499 Tetraspanin/Peripherin
Gene 3D
CDD
ProteinModelPortal
PDB
5JA4
E-value=1.868e-30,
Score=336
Ontologies
KEGG
GO
Topology
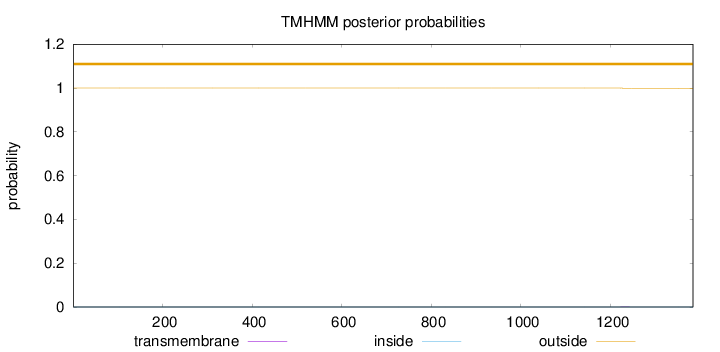
Length:
1384
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00974
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.00007
outside
1 - 1384
Population Genetic Test Statistics
Pi
22.992818
Theta
23.483132
Tajima's D
0.040577
CLR
1.345477
CSRT
0.381480925953702
Interpretation
Uncertain
