Gene
KWMTBOMO06833
Pre Gene Modal
BGIBMGA012094
Annotation
PREDICTED:_tyrosine-protein_kinase_Src64B_isoform_X1_[Bombyx_mori]
Full name
Tyrosine-protein kinase
+ More
Tyrosine-protein kinase Src64B
Tyrosine-protein kinase Src64B
Location in the cell
Cytoplasmic Reliability : 2.536
Sequence
CDS
ATGGGTAACAAGTGTTGCAGCCGCCGACATGACGCCGAGAGACCTCCTGTGTACCCTCACAAGAATGAGCCCGGTTTCACAACGTGCCCCGTGTCTTTTGACACCCGGTACACTGGCGAGCCAAACACCAGGCAGCCGTCGCGACCTCCTGTTGACGTCGTAAGAACGACAACAAAATGCATATCAAACGGCGGTCGTCGAGTCGTAAAAGCCCTATGCGAGTATACGGCTCGAGCGGACTCCGACATCTCGTTTCACAAGGGCGACCGCATGGAAGTCCTCAACAGCTCGGAGCCGGATTGGTGGATGGTGTTCCATCTGACCACGAGGAGGGAAGGCCTGGTTCCAGTCAATTACGTCGCCGAAGAGAGTTCAGTGGAATGTGAAGACTGGTTCTTTCCTCACGTGTCGAGGAAGGAGGCCGACAAACTCCTCCTAGCCGAGAGCAACCCCCGCGGTACGTTCTTGGTCCGACCGGCGGAGCACAACCCTCACGGCTTCAGCTTAAGCGTCAAAGATTGGGAAGACGGAAGGGGCTACCACGTGAAACATTATAAAATAAAGCCGCTCGACAAAGGCGGTTTTTATATCGCCACAAACCAAACCTTCCCGAGCTTACCCGCTCTCGTCATGTCGTATACGAAAAATGCATTAGGGCTGTGTCACGTCCTGGCCCGGCCTTGTCCGAAGCCCGAGCCGCAGATCTGGGACCTCGGGCCCGAACTGCGGGACAAGTGGGAGATACCCAGGATGGAAATACAACTCATTAGGAAGCTCGGTCAAGGAAACTTCGGCGAAGTCTATTACGGGAAATGGAGGAATAATATTGAGGTCGCCGTGAAGACTCTTCGCGAGGGCACAATGTCGACTCAGGCCTTCCTGCAAGAGGCGGCGATCATGAAGAAGTTCCGGCACAACAAGCTGGTCGCCCTGTACGCCGTCTGCTCGCAGCAGGAGCCGGTCTACATCGTCCAGGAGTACATGAGCCGCGGCTCCTTACTGGAGTTCTTGAGGAACGGCGAAGGGAAGTTCCTCCAGTTCGAGGACCTAGTTTACATAGCGGCCCAGGTCGCCTCCGGTATGGAGTATCTCGAATCGAAAATGCTGATTCACAGAGACCTGGCCGCTCGGAACGTGCTCATCGGGGAGAACAATACCGCCAAAATATGCGACTTCGGTCTCGCCAGGGTCATAGAAGACAACGAATACTGCCCCAAGCAAGGCTCTAGGTTCCCAGTCAAATGGACAGCCCCCGAAGCTATAATATACGGGAAGTTCTCCATCAAAAGCGACGTCTGGTCGTACGGCATTCTCTTAATGGAGCTTTTCACATACGGGAAGATTCCGTACCCGGGATTGCACGGCAAGGACGTCATCGACAAAATCGAGATCGGCTACCGGATGCCGAAGCCGATAGACGTGTACCTGCCGGACGCCATATACGGGCTGATGCTGCAGTGCTGGGACGCGGTGCCTGAGAAGCGGCCCACCTTCGAGTTCCTCAATCATTACTTCGAATCGTTTACGGTGACCAGCGAGATGCCGTACAAGGAGGTACACGATTGA
Protein
MGNKCCSRRHDAERPPVYPHKNEPGFTTCPVSFDTRYTGEPNTRQPSRPPVDVVRTTTKCISNGGRRVVKALCEYTARADSDISFHKGDRMEVLNSSEPDWWMVFHLTTRREGLVPVNYVAEESSVECEDWFFPHVSRKEADKLLLAESNPRGTFLVRPAEHNPHGFSLSVKDWEDGRGYHVKHYKIKPLDKGGFYIATNQTFPSLPALVMSYTKNALGLCHVLARPCPKPEPQIWDLGPELRDKWEIPRMEIQLIRKLGQGNFGEVYYGKWRNNIEVAVKTLREGTMSTQAFLQEAAIMKKFRHNKLVALYAVCSQQEPVYIVQEYMSRGSLLEFLRNGEGKFLQFEDLVYIAAQVASGMEYLESKMLIHRDLAARNVLIGENNTAKICDFGLARVIEDNEYCPKQGSRFPVKWTAPEAIIYGKFSIKSDVWSYGILLMELFTYGKIPYPGLHGKDVIDKIEIGYRMPKPIDVYLPDAIYGLMLQCWDAVPEKRPTFEFLNHYFESFTVTSEMPYKEVHD
Summary
Description
May play a role in the development of neural tissue and smooth muscle.
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the protein kinase superfamily. Tyr protein kinase family. SRC subfamily.
Belongs to the protein kinase superfamily. Tyr protein kinase family. SRC subfamily.
Keywords
ATP-binding
Complete proteome
Developmental protein
Kinase
Nucleotide-binding
Phosphoprotein
Proto-oncogene
Reference proteome
SH2 domain
SH3 domain
Transferase
Tyrosine-protein kinase
Feature
chain Tyrosine-protein kinase Src64B
Uniprot
A0A2A4JUW5
A0A2A4JVG0
H9JRD4
A0A212ERZ7
A0A194PEH4
A0A194R3F6
+ More
A0A0L7L919 A0A084WMI0 W5JAG1 A0A1Q3G073 A0A182MD62 A0A182QU87 A0A0K8VLV5 W8B4I4 A0A034V3V1 A0A182V1T8 A0A182UFX5 Q7Q5D3 A0A182J610 Q17CE8 A0A0A1WLL3 A0A1I8MNN6 A0A1J1IN93 A0A1S4F874 A0A3B0JC04 Q29EB5 B4H1S6 A0A1W4VNP7 A0A1I8Q8J8 B4HU86 B3NC61 B4QR05 M9MSK3 B4PI98 P00528 A0A0Q9VZD8 B4MFY6 B4IZ80 A0A0R3P952 B4KX98 A0A1A9X352 A0A1A9UXP5 A0A1A9XXA5 B4MMY8 A0A1A9ZWJ9 A0A1W4VAL7 X2JCI2 B3M9I4 A0A1Q3G0A3 A0A3B0JXY4 A0A3B0JQ36 A0A182GE84 A0A0C9S1N2 A0A0C9RZ52 E2B6N7 A0A182T948 A0A195E5N9 F4W469 A0A151X1X9 E2AGB2 A0A1B6DBV7 A0A310S576 A0A2J7QBD3 A0A158NF94 A0A195CLJ5 A0A2J7QBD7 A0A195ET97 A0A0C9QIX9 A0A026W269 A0A3L8DPV5 A0A0L7QSY0 A0A1Y1MNH6 A0A067RF36 A0A1B0AZ14 A0A0N0BIX5 E9I9W3 A0A2A3EBZ5 A0A195B436 A0A154PDP5 A0A1B6F6I2 A0A2S2QXI1 A0A2H8TTJ2 J9JNW3 A0A224XAP3 A0A2S2NKV7 D6WH54 A0A023F4P5 T1I1G4 A0A088A505 A0A0J7L1N8 A0A0A9W1P4 A0A1B6M227 A0A0T6B3B8 E0VM96 B0WKZ9 A0A1D2MCK7 A0A0N8DE06 A0A0N8AZW3 A0A0P5WRF2 A0A0P5X2C5
A0A0L7L919 A0A084WMI0 W5JAG1 A0A1Q3G073 A0A182MD62 A0A182QU87 A0A0K8VLV5 W8B4I4 A0A034V3V1 A0A182V1T8 A0A182UFX5 Q7Q5D3 A0A182J610 Q17CE8 A0A0A1WLL3 A0A1I8MNN6 A0A1J1IN93 A0A1S4F874 A0A3B0JC04 Q29EB5 B4H1S6 A0A1W4VNP7 A0A1I8Q8J8 B4HU86 B3NC61 B4QR05 M9MSK3 B4PI98 P00528 A0A0Q9VZD8 B4MFY6 B4IZ80 A0A0R3P952 B4KX98 A0A1A9X352 A0A1A9UXP5 A0A1A9XXA5 B4MMY8 A0A1A9ZWJ9 A0A1W4VAL7 X2JCI2 B3M9I4 A0A1Q3G0A3 A0A3B0JXY4 A0A3B0JQ36 A0A182GE84 A0A0C9S1N2 A0A0C9RZ52 E2B6N7 A0A182T948 A0A195E5N9 F4W469 A0A151X1X9 E2AGB2 A0A1B6DBV7 A0A310S576 A0A2J7QBD3 A0A158NF94 A0A195CLJ5 A0A2J7QBD7 A0A195ET97 A0A0C9QIX9 A0A026W269 A0A3L8DPV5 A0A0L7QSY0 A0A1Y1MNH6 A0A067RF36 A0A1B0AZ14 A0A0N0BIX5 E9I9W3 A0A2A3EBZ5 A0A195B436 A0A154PDP5 A0A1B6F6I2 A0A2S2QXI1 A0A2H8TTJ2 J9JNW3 A0A224XAP3 A0A2S2NKV7 D6WH54 A0A023F4P5 T1I1G4 A0A088A505 A0A0J7L1N8 A0A0A9W1P4 A0A1B6M227 A0A0T6B3B8 E0VM96 B0WKZ9 A0A1D2MCK7 A0A0N8DE06 A0A0N8AZW3 A0A0P5WRF2 A0A0P5X2C5
EC Number
2.7.10.2
Pubmed
19121390
22118469
26354079
26227816
24438588
20920257
+ More
23761445 24495485 25348373 12364791 14747013 17210077 17510324 25830018 25315136 15632085 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 2996778 12537569 6317185 9731193 18057021 26483478 20798317 21719571 21347285 24508170 30249741 28004739 24845553 21282665 18362917 19820115 25474469 25401762 20566863 27289101
23761445 24495485 25348373 12364791 14747013 17210077 17510324 25830018 25315136 15632085 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 2996778 12537569 6317185 9731193 18057021 26483478 20798317 21719571 21347285 24508170 30249741 28004739 24845553 21282665 18362917 19820115 25474469 25401762 20566863 27289101
EMBL
NWSH01000537
PCG75801.1
PCG75799.1
BABH01016221
BABH01016222
AGBW02012898
+ More
OWR44255.1 KQ459606 KPI91756.1 KQ460779 KPJ12338.1 JTDY01002220 KOB71865.1 ATLV01024459 KE525352 KFB51424.1 ADMH02001836 ETN60976.1 GFDL01001834 JAV33211.1 AXCM01008822 AXCN02000463 GDHF01032063 GDHF01025214 GDHF01018527 GDHF01012486 JAI20251.1 JAI27100.1 JAI33787.1 JAI39828.1 GAMC01018351 GAMC01018350 GAMC01018349 JAB88206.1 GAKP01022133 GAKP01022131 GAKP01022130 GAKP01022129 JAC36822.1 AAAB01008960 EAA11388.2 CH477309 EAT44003.1 GBXI01014358 GBXI01005471 JAC99933.1 JAD08821.1 CVRI01000056 CRL01691.1 OUUW01000002 SPP77582.1 CH379070 EAL30146.1 CH479203 EDW30278.1 CH480817 EDW50507.1 CH954178 EDV50949.1 CM000363 CM002912 EDX09242.1 KMY97648.1 AE014296 ADV37488.1 CM000159 EDW93440.1 M11917 AY051781 K01043 AJ002919 CH940669 KRF78195.1 EDW58247.1 CH916366 CH916865 EDV95602.1 EDW04576.1 KRT08678.1 CH933809 EDW17556.1 CH963847 EDW73544.1 AHN57962.1 CH902618 EDV38990.1 KPU77803.1 GFDL01001835 JAV33210.1 SPP77581.1 SPP77580.1 JXUM01057599 JXUM01057600 KQ561962 KXJ77052.1 GBYB01014691 JAG84458.1 GBYB01014690 JAG84457.1 GL446000 EFN88621.1 KQ979609 KYN20164.1 GL887491 EGI71065.1 KQ982585 KYQ54295.1 GL439266 EFN67603.1 GEDC01016649 GEDC01014176 JAS20649.1 JAS23122.1 KQ771134 OAD52547.1 NEVH01016301 PNF25896.1 ADTU01001526 ADTU01001527 KQ977642 KYN00944.1 PNF25895.1 KQ981979 KYN31475.1 GBYB01014693 JAG84460.1 KK107538 EZA49129.1 QOIP01000005 RLU22454.1 KQ414755 KOC61730.1 GEZM01029673 GEZM01029672 JAV85995.1 KK852729 KDR17588.1 JXJN01006116 KQ435727 KOX78091.1 GL761846 EFZ22631.1 KZ288303 PBC28812.1 KQ976625 KYM79027.1 KQ434870 KZC09514.1 GECZ01030960 GECZ01023941 GECZ01023642 GECZ01014541 JAS38809.1 JAS45828.1 JAS46127.1 JAS55228.1 GGMS01013017 MBY82220.1 GFXV01002789 GFXV01005741 MBW14594.1 MBW17546.1 ABLF02040183 GFTR01006969 JAW09457.1 GGMR01005129 MBY17748.1 KQ971330 EEZ99753.2 GBBI01002330 JAC16382.1 ACPB03006877 LBMM01001290 KMQ96571.1 GBHO01041871 GBHO01041870 JAG01733.1 JAG01734.1 GEBQ01012640 GEBQ01009978 GEBQ01002389 JAT27337.1 JAT29999.1 JAT37588.1 LJIG01016022 KRT81848.1 DS235303 EEB14502.1 DS231978 EDS30139.1 LJIJ01001816 ODM90737.1 GDIP01042999 LRGB01001581 JAM60716.1 KZS11042.1 GDIQ01226555 GDIQ01036940 JAK25170.1 GDIP01083298 JAM20417.1 GDIP01078849 JAM24866.1
OWR44255.1 KQ459606 KPI91756.1 KQ460779 KPJ12338.1 JTDY01002220 KOB71865.1 ATLV01024459 KE525352 KFB51424.1 ADMH02001836 ETN60976.1 GFDL01001834 JAV33211.1 AXCM01008822 AXCN02000463 GDHF01032063 GDHF01025214 GDHF01018527 GDHF01012486 JAI20251.1 JAI27100.1 JAI33787.1 JAI39828.1 GAMC01018351 GAMC01018350 GAMC01018349 JAB88206.1 GAKP01022133 GAKP01022131 GAKP01022130 GAKP01022129 JAC36822.1 AAAB01008960 EAA11388.2 CH477309 EAT44003.1 GBXI01014358 GBXI01005471 JAC99933.1 JAD08821.1 CVRI01000056 CRL01691.1 OUUW01000002 SPP77582.1 CH379070 EAL30146.1 CH479203 EDW30278.1 CH480817 EDW50507.1 CH954178 EDV50949.1 CM000363 CM002912 EDX09242.1 KMY97648.1 AE014296 ADV37488.1 CM000159 EDW93440.1 M11917 AY051781 K01043 AJ002919 CH940669 KRF78195.1 EDW58247.1 CH916366 CH916865 EDV95602.1 EDW04576.1 KRT08678.1 CH933809 EDW17556.1 CH963847 EDW73544.1 AHN57962.1 CH902618 EDV38990.1 KPU77803.1 GFDL01001835 JAV33210.1 SPP77581.1 SPP77580.1 JXUM01057599 JXUM01057600 KQ561962 KXJ77052.1 GBYB01014691 JAG84458.1 GBYB01014690 JAG84457.1 GL446000 EFN88621.1 KQ979609 KYN20164.1 GL887491 EGI71065.1 KQ982585 KYQ54295.1 GL439266 EFN67603.1 GEDC01016649 GEDC01014176 JAS20649.1 JAS23122.1 KQ771134 OAD52547.1 NEVH01016301 PNF25896.1 ADTU01001526 ADTU01001527 KQ977642 KYN00944.1 PNF25895.1 KQ981979 KYN31475.1 GBYB01014693 JAG84460.1 KK107538 EZA49129.1 QOIP01000005 RLU22454.1 KQ414755 KOC61730.1 GEZM01029673 GEZM01029672 JAV85995.1 KK852729 KDR17588.1 JXJN01006116 KQ435727 KOX78091.1 GL761846 EFZ22631.1 KZ288303 PBC28812.1 KQ976625 KYM79027.1 KQ434870 KZC09514.1 GECZ01030960 GECZ01023941 GECZ01023642 GECZ01014541 JAS38809.1 JAS45828.1 JAS46127.1 JAS55228.1 GGMS01013017 MBY82220.1 GFXV01002789 GFXV01005741 MBW14594.1 MBW17546.1 ABLF02040183 GFTR01006969 JAW09457.1 GGMR01005129 MBY17748.1 KQ971330 EEZ99753.2 GBBI01002330 JAC16382.1 ACPB03006877 LBMM01001290 KMQ96571.1 GBHO01041871 GBHO01041870 JAG01733.1 JAG01734.1 GEBQ01012640 GEBQ01009978 GEBQ01002389 JAT27337.1 JAT29999.1 JAT37588.1 LJIG01016022 KRT81848.1 DS235303 EEB14502.1 DS231978 EDS30139.1 LJIJ01001816 ODM90737.1 GDIP01042999 LRGB01001581 JAM60716.1 KZS11042.1 GDIQ01226555 GDIQ01036940 JAK25170.1 GDIP01083298 JAM20417.1 GDIP01078849 JAM24866.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000030765 UP000000673 UP000075883 UP000075886 UP000075903 UP000075902 UP000007062 UP000075880 UP000008820 UP000095301 UP000183832 UP000268350 UP000001819 UP000008744 UP000192221 UP000095300 UP000001292 UP000008711 UP000000304 UP000000803 UP000002282 UP000008792 UP000001070 UP000009192 UP000091820 UP000078200 UP000092443 UP000007798 UP000092445 UP000007801 UP000069940 UP000249989 UP000008237 UP000075901 UP000078492 UP000007755 UP000075809 UP000000311 UP000235965 UP000005205 UP000078542 UP000078541 UP000053097 UP000279307 UP000053825 UP000027135 UP000092460 UP000053105 UP000242457 UP000078540 UP000076502 UP000007819 UP000007266 UP000015103 UP000005203 UP000036403 UP000009046 UP000002320 UP000094527 UP000076858
UP000030765 UP000000673 UP000075883 UP000075886 UP000075903 UP000075902 UP000007062 UP000075880 UP000008820 UP000095301 UP000183832 UP000268350 UP000001819 UP000008744 UP000192221 UP000095300 UP000001292 UP000008711 UP000000304 UP000000803 UP000002282 UP000008792 UP000001070 UP000009192 UP000091820 UP000078200 UP000092443 UP000007798 UP000092445 UP000007801 UP000069940 UP000249989 UP000008237 UP000075901 UP000078492 UP000007755 UP000075809 UP000000311 UP000235965 UP000005205 UP000078542 UP000078541 UP000053097 UP000279307 UP000053825 UP000027135 UP000092460 UP000053105 UP000242457 UP000078540 UP000076502 UP000007819 UP000007266 UP000015103 UP000005203 UP000036403 UP000009046 UP000002320 UP000094527 UP000076858
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JUW5
A0A2A4JVG0
H9JRD4
A0A212ERZ7
A0A194PEH4
A0A194R3F6
+ More
A0A0L7L919 A0A084WMI0 W5JAG1 A0A1Q3G073 A0A182MD62 A0A182QU87 A0A0K8VLV5 W8B4I4 A0A034V3V1 A0A182V1T8 A0A182UFX5 Q7Q5D3 A0A182J610 Q17CE8 A0A0A1WLL3 A0A1I8MNN6 A0A1J1IN93 A0A1S4F874 A0A3B0JC04 Q29EB5 B4H1S6 A0A1W4VNP7 A0A1I8Q8J8 B4HU86 B3NC61 B4QR05 M9MSK3 B4PI98 P00528 A0A0Q9VZD8 B4MFY6 B4IZ80 A0A0R3P952 B4KX98 A0A1A9X352 A0A1A9UXP5 A0A1A9XXA5 B4MMY8 A0A1A9ZWJ9 A0A1W4VAL7 X2JCI2 B3M9I4 A0A1Q3G0A3 A0A3B0JXY4 A0A3B0JQ36 A0A182GE84 A0A0C9S1N2 A0A0C9RZ52 E2B6N7 A0A182T948 A0A195E5N9 F4W469 A0A151X1X9 E2AGB2 A0A1B6DBV7 A0A310S576 A0A2J7QBD3 A0A158NF94 A0A195CLJ5 A0A2J7QBD7 A0A195ET97 A0A0C9QIX9 A0A026W269 A0A3L8DPV5 A0A0L7QSY0 A0A1Y1MNH6 A0A067RF36 A0A1B0AZ14 A0A0N0BIX5 E9I9W3 A0A2A3EBZ5 A0A195B436 A0A154PDP5 A0A1B6F6I2 A0A2S2QXI1 A0A2H8TTJ2 J9JNW3 A0A224XAP3 A0A2S2NKV7 D6WH54 A0A023F4P5 T1I1G4 A0A088A505 A0A0J7L1N8 A0A0A9W1P4 A0A1B6M227 A0A0T6B3B8 E0VM96 B0WKZ9 A0A1D2MCK7 A0A0N8DE06 A0A0N8AZW3 A0A0P5WRF2 A0A0P5X2C5
A0A0L7L919 A0A084WMI0 W5JAG1 A0A1Q3G073 A0A182MD62 A0A182QU87 A0A0K8VLV5 W8B4I4 A0A034V3V1 A0A182V1T8 A0A182UFX5 Q7Q5D3 A0A182J610 Q17CE8 A0A0A1WLL3 A0A1I8MNN6 A0A1J1IN93 A0A1S4F874 A0A3B0JC04 Q29EB5 B4H1S6 A0A1W4VNP7 A0A1I8Q8J8 B4HU86 B3NC61 B4QR05 M9MSK3 B4PI98 P00528 A0A0Q9VZD8 B4MFY6 B4IZ80 A0A0R3P952 B4KX98 A0A1A9X352 A0A1A9UXP5 A0A1A9XXA5 B4MMY8 A0A1A9ZWJ9 A0A1W4VAL7 X2JCI2 B3M9I4 A0A1Q3G0A3 A0A3B0JXY4 A0A3B0JQ36 A0A182GE84 A0A0C9S1N2 A0A0C9RZ52 E2B6N7 A0A182T948 A0A195E5N9 F4W469 A0A151X1X9 E2AGB2 A0A1B6DBV7 A0A310S576 A0A2J7QBD3 A0A158NF94 A0A195CLJ5 A0A2J7QBD7 A0A195ET97 A0A0C9QIX9 A0A026W269 A0A3L8DPV5 A0A0L7QSY0 A0A1Y1MNH6 A0A067RF36 A0A1B0AZ14 A0A0N0BIX5 E9I9W3 A0A2A3EBZ5 A0A195B436 A0A154PDP5 A0A1B6F6I2 A0A2S2QXI1 A0A2H8TTJ2 J9JNW3 A0A224XAP3 A0A2S2NKV7 D6WH54 A0A023F4P5 T1I1G4 A0A088A505 A0A0J7L1N8 A0A0A9W1P4 A0A1B6M227 A0A0T6B3B8 E0VM96 B0WKZ9 A0A1D2MCK7 A0A0N8DE06 A0A0N8AZW3 A0A0P5WRF2 A0A0P5X2C5
PDB
2H8H
E-value=1.71532e-143,
Score=1307
Ontologies
KEGG
PATHWAY
GO
GO:0005524
GO:0004715
GO:0030154
GO:0031234
GO:0005102
GO:0038083
GO:0007169
GO:0018108
GO:0036058
GO:0090136
GO:0005886
GO:0048167
GO:0007349
GO:0007300
GO:0030708
GO:0030717
GO:0007301
GO:0007616
GO:0007411
GO:0008302
GO:0007293
GO:0048477
GO:0034332
GO:0045743
GO:0016319
GO:0005829
GO:0007435
GO:0030036
GO:0045742
GO:0120176
GO:0045172
GO:0030723
GO:0008064
GO:0007424
GO:0008335
GO:0030046
GO:0045886
GO:0000185
GO:0045874
GO:0004713
GO:0004672
GO:0006468
GO:0005515
GO:0043565
GO:0016567
GO:0016020
GO:0005634
GO:0006260
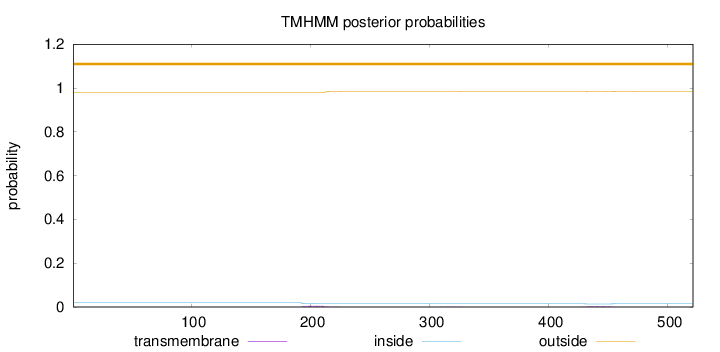
Topology
Length:
521
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16682
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01994
outside
1 - 521
Population Genetic Test Statistics
Pi
19.15826
Theta
16.841685
Tajima's D
-1.330197
CLR
1.119516
CSRT
0.0808959552022399
Interpretation
Uncertain
