Gene
KWMTBOMO06832
Pre Gene Modal
BGIBMGA012057
Annotation
PREDICTED:_solute_carrier_family_35_member_B3_isoform_X1_[Bombyx_mori]
Full name
Adenosine 3'-phospho 5'-phosphosulfate transporter 2
Alternative Name
PAPS transporter 2
Solute carrier family 35 member B3 homolog
dPAPST2
Solute carrier family 35 member B3
Solute carrier family 35 member B3 homolog
dPAPST2
Solute carrier family 35 member B3
Location in the cell
PlasmaMembrane Reliability : 4.954
Sequence
CDS
ATGGAAACGTCATCAGATAAAATGAATTCTGTGAAGGAAACCTTCATAAGGATAGATGATGAGAACAGCAGAACATCAAAAACAGAAATCAATATATTATGCTTTGACATAACAAATTACAGTCAATTAACTCAGTTCCTGTGTTGTTCGATATTCGTTTTTATATTCTATTTAGCTTATGGATACTTCTTAGAATTAATATTTGCAAAACCGGAAGTGAAGCCAGTCAGTTTGTATATAACCTTAGTGCAATTTATGATTACTATGTTGTTGAGTTATGGAGAATCATGGATAAGAAATCCTATCAAAAGGAAAGTTCCACTAAAAACGTATGCAGTTTTAGCTGCATTGACATTAGGAACAATGTCATTTTCGAATTTAGCTTTAAGTTATTTGAACTATCCAACTCAATTGATATTCAAGAGCTGTAAACTTATACCAGTTATGATTGGTAGTATTATCATAATGAGAAAACGTTATTCGTTTTTGGATTATGTAGCGGCAATTGTTATGTGTGTAGGACTGACTATGTTTACTCTGGCTGACTCCAGCACATCACCGAATTTCGATTTGATTGGCGTTCTAGTTATATCCTTAGCATTACTATGCGACGCAATAATCGGAAACGTACAAGAGAAAGCAATGAAACAGTACCAAGCTTCAAATAACGAAGTTGTTTTCTATTCATATGCGATTGCATGTGTTTACCTTGTCTGTATTACTGGATTCAGTGGTATACTAGTTGACGGATTTGCTTATTGTGCTGAGACACCAGTTGAAATGTACCGGAACATATTTCTTCTAAGTCTGAGCGGCTATATGGGTCTGCAAGCGGTGTTGACGCTGGTGAGGATATGCGGTGCGACGGTCGCCGTTACCGTCACCACGATGAGGAAAGCTCTCTCGATAATCATATCGTTCTTGTTGTTCAGCAAGCCTTTTGTCTTCCAGTACGTTTGGTCGGGATCTTTGGTGGTTTTAGCCATTTACATAAATCACTACAGTAAAAAGCATCCCCACTACGTACCGGCCTCACTGTCTAGTTGCTGGCAACAAATAAGGAACTATTACGATTCAGAATACAGATATCTTAAGATGAAAACTAAAATGTACACCGATACGGTGTAA
Protein
METSSDKMNSVKETFIRIDDENSRTSKTEINILCFDITNYSQLTQFLCCSIFVFIFYLAYGYFLELIFAKPEVKPVSLYITLVQFMITMLLSYGESWIRNPIKRKVPLKTYAVLAALTLGTMSFSNLALSYLNYPTQLIFKSCKLIPVMIGSIIIMRKRYSFLDYVAAIVMCVGLTMFTLADSSTSPNFDLIGVLVISLALLCDAIIGNVQEKAMKQYQASNNEVVFYSYAIACVYLVCITGFSGILVDGFAYCAETPVEMYRNIFLLSLSGYMGLQAVLTLVRICGATVAVTVTTMRKALSIIISFLLFSKPFVFQYVWSGSLVVLAIYINHYSKKHPHYVPASLSSCWQQIRNYYDSEYRYLKMKTKMYTDTV
Summary
Description
Mediates the transport of adenosine 3'-phospho 5'-phosphosulfate (PAPS), from cytosol into Golgi. PAPS is a universal sulfuryl donor for sulfation events that take place in the Golgi. Essential for viability. Involved in glycosaminoglycan synthesis and the subsequent signaling. May be involved in hh and dpp signaling by controlling the sulfation of heparan sulfate (HS) (By similarity).
Mediates the transport of adenosine 3'-phospho 5'-phosphosulfate (PAPS), from cytosol into Golgi. PAPS is a universal sulfuryl donor for sulfation events that take place in the Golgi. Essential for viability. Involved in glycosaminoglycan synthesis and the subsequent signaling. May be involved in hh and dpp signaling by controlling the sulfation of heparan sulfate (HS).
Mediates the transport of adenosine 3'-phospho 5'-phosphosulfate (PAPS), from cytosol into Golgi. PAPS is a universal sulfuryl donor for sulfation events that take place in the Golgi. Compensates for the insufficient expression of SLC35B2/PAPST1 during the synthesis of sulfated glycoconjugates in the colon (By similarity).
Mediates the transport of adenosine 3'-phospho 5'-phosphosulfate (PAPS), from cytosol into Golgi. PAPS is a universal sulfuryl donor for sulfation events that take place in the Golgi. Essential for viability. Involved in glycosaminoglycan synthesis and the subsequent signaling. May be involved in hh and dpp signaling by controlling the sulfation of heparan sulfate (HS).
Mediates the transport of adenosine 3'-phospho 5'-phosphosulfate (PAPS), from cytosol into Golgi. PAPS is a universal sulfuryl donor for sulfation events that take place in the Golgi. Compensates for the insufficient expression of SLC35B2/PAPST1 during the synthesis of sulfated glycoconjugates in the colon (By similarity).
Biophysicochemical Properties
2.3 uM for PAPS
Similarity
Belongs to the nucleotide-sugar transporter family. SLC35B subfamily.
Keywords
Complete proteome
Developmental protein
Golgi apparatus
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Glycoprotein
Phosphoprotein
Feature
chain Adenosine 3'-phospho 5'-phosphosulfate transporter 2
Uniprot
D2XRA6
Q2F658
A0A2A4JWN3
A0A3S2NGP5
H9JR97
A0A1E1WA90
+ More
A0A212ERX1 A0A0L7L8N2 A0A194R3S0 A0A194PKX3 A0A0K8TQS0 A0A1Y1LDF0 A0A232FGK7 D6WH53 A0A2M4AKV7 K7J9A9 N6T0M2 A0A084WMH9 A0A2M4BRL8 A0A2M3Z4X2 W5J947 A0A1Q3FBK9 A0A2M4BRH8 A0A182NIY5 B0WKZ8 U4U9X4 A0A182ILV1 Q7Q5D4 A0A182URW5 A0A023ETA5 A0A182X956 A0A182I493 A0A182GE83 B4PJY7 A0A182TGT5 B4LBF8 Q9VVD9 A0A1W4VSK1 Q17CE7 A0A182QTK9 B4QNE6 A0A182RR65 A0A1B0B7S8 B3NDD6 A0A182MU49 B4HKA6 A0A182Y7V4 B4H5P1 A0A0R3P5S5 Q29EY2 A0A182WBY4 A0A1B6JGM3 A0A034V9Y8 B3M5H5 A0A182SJS2 A0A182K706 A0A3B0KFW5 B4N4X0 A0A1D2N287 A0A0K8VMZ2 W8BKZ8 A0A182NZT2 B4J0G6 R7UJB8 A0A0V0GBN0 A0A0L0CL30 A0A1I8MAJ6 A0A0A1X117 A0A1I8P623 A0A069DRV6 B4KV26 A0A1B6KRQ8 A0A2J7QBC8 A0A2S2P8I9 A0A164P2R6 A0A2R7WJU4 A0A0P6I6Q3 A0A2C9LTE5 E9H620 T1HPW5 A0A067R5L8 A0A2H8TPM1 A0A1A9VMZ9 A0A0P6AHT6 A0A1B0A2J2 A0A0B6ZVJ6 A0A1S3H0C6 A0A099ZIK7 A0A3P8VQG3 W5N6L6 A0A3B5KLF8 A0A1S3J8P5 A0A091I7L1 D4AAG3 Q922Q5 A0A146LSY8 A0A091MGF2 A0A2D0PQQ4 A0A0A9YSM8
A0A212ERX1 A0A0L7L8N2 A0A194R3S0 A0A194PKX3 A0A0K8TQS0 A0A1Y1LDF0 A0A232FGK7 D6WH53 A0A2M4AKV7 K7J9A9 N6T0M2 A0A084WMH9 A0A2M4BRL8 A0A2M3Z4X2 W5J947 A0A1Q3FBK9 A0A2M4BRH8 A0A182NIY5 B0WKZ8 U4U9X4 A0A182ILV1 Q7Q5D4 A0A182URW5 A0A023ETA5 A0A182X956 A0A182I493 A0A182GE83 B4PJY7 A0A182TGT5 B4LBF8 Q9VVD9 A0A1W4VSK1 Q17CE7 A0A182QTK9 B4QNE6 A0A182RR65 A0A1B0B7S8 B3NDD6 A0A182MU49 B4HKA6 A0A182Y7V4 B4H5P1 A0A0R3P5S5 Q29EY2 A0A182WBY4 A0A1B6JGM3 A0A034V9Y8 B3M5H5 A0A182SJS2 A0A182K706 A0A3B0KFW5 B4N4X0 A0A1D2N287 A0A0K8VMZ2 W8BKZ8 A0A182NZT2 B4J0G6 R7UJB8 A0A0V0GBN0 A0A0L0CL30 A0A1I8MAJ6 A0A0A1X117 A0A1I8P623 A0A069DRV6 B4KV26 A0A1B6KRQ8 A0A2J7QBC8 A0A2S2P8I9 A0A164P2R6 A0A2R7WJU4 A0A0P6I6Q3 A0A2C9LTE5 E9H620 T1HPW5 A0A067R5L8 A0A2H8TPM1 A0A1A9VMZ9 A0A0P6AHT6 A0A1B0A2J2 A0A0B6ZVJ6 A0A1S3H0C6 A0A099ZIK7 A0A3P8VQG3 W5N6L6 A0A3B5KLF8 A0A1S3J8P5 A0A091I7L1 D4AAG3 Q922Q5 A0A146LSY8 A0A091MGF2 A0A2D0PQQ4 A0A0A9YSM8
Pubmed
19121390
22118469
26227816
26354079
26369729
28004739
+ More
28648823 18362917 19820115 20075255 23537049 24438588 20920257 23761445 12364791 24945155 26483478 17994087 17550304 16873373 10731132 12537572 12537569 18327897 17510324 22936249 25244985 15632085 25348373 27289101 24495485 23254933 26108605 25315136 25830018 26334808 15562597 21292972 24845553 24487278 21551351 15057822 16141072 15489334 26823975 25401762
28648823 18362917 19820115 20075255 23537049 24438588 20920257 23761445 12364791 24945155 26483478 17994087 17550304 16873373 10731132 12537572 12537569 18327897 17510324 22936249 25244985 15632085 25348373 27289101 24495485 23254933 26108605 25315136 25830018 26334808 15562597 21292972 24845553 24487278 21551351 15057822 16141072 15489334 26823975 25401762
EMBL
GU244352
ADB08385.1
DQ311214
ABD36159.1
NWSH01000537
PCG75802.1
+ More
RSAL01000045 RVE50624.1 BABH01016222 GDQN01007142 JAT83912.1 AGBW02012898 OWR44256.1 JTDY01002220 KOB71867.1 KQ460779 KPJ12337.1 KQ459606 KPI91755.1 GDAI01000899 JAI16704.1 GEZM01061560 JAV70370.1 NNAY01000233 OXU29825.1 KQ971330 EFA00972.1 GGFK01008041 MBW41362.1 APGK01049756 KB741156 ENN73619.1 ATLV01024459 KE525352 KFB51423.1 GGFJ01006482 MBW55623.1 GGFM01002794 MBW23545.1 ADMH02001836 ETN60977.1 GFDL01010081 JAV24964.1 GGFJ01006481 MBW55622.1 DS231978 EDS30138.1 KB631941 ERL87376.1 AAAB01008960 GAPW01001984 JAC11614.1 APCN01002286 JXUM01057598 KQ561962 KXJ77051.1 CM000159 EDW94756.2 CH940647 EDW69746.1 AB242867 AE014296 AY069640 CH477309 EAT44004.1 AXCN02000463 CM000363 CM002912 EDX10816.1 KMZ00187.1 JXJN01009712 CH954178 EDV51998.2 AXCM01003551 CH480815 EDW41847.1 CH479211 EDW33093.1 CH379070 KRT08497.1 GECU01009367 JAS98339.1 GAKP01020367 JAC38585.1 CH902618 EDV39585.1 OUUW01000009 SPP85219.1 CH964101 EDW79409.1 LJIJ01000285 ODM99368.1 GDHF01012065 JAI40249.1 GAMC01007118 JAB99437.1 CH916366 EDV96802.1 AMQN01008493 KB303236 ELU03367.1 GECL01000832 JAP05292.1 JRES01000254 KNC32956.1 GBXI01009696 JAD04596.1 GBGD01002086 JAC86803.1 CH933809 EDW18337.2 GEBQ01026053 JAT13924.1 NEVH01016301 PNF25893.1 GGMR01012889 MBY25508.1 LRGB01002722 KZS06460.1 KK854954 PTY19914.1 GDIQ01008627 JAN86110.1 GL732595 EFX72822.1 ACPB03001903 KK852729 KDR17582.1 GFXV01004135 MBW15940.1 GDIP01034394 JAM69321.1 HACG01025774 CEK72639.1 KL894251 KGL81647.1 AHAT01002330 KL218247 KFP03433.1 AABR07027364 AK076592 AK077208 AK141280 BC006881 GDHC01009079 JAQ09550.1 KK827143 KFP73587.1 GBHO01008400 GBHO01001380 GBHO01001379 JAG35204.1 JAG42224.1 JAG42225.1
RSAL01000045 RVE50624.1 BABH01016222 GDQN01007142 JAT83912.1 AGBW02012898 OWR44256.1 JTDY01002220 KOB71867.1 KQ460779 KPJ12337.1 KQ459606 KPI91755.1 GDAI01000899 JAI16704.1 GEZM01061560 JAV70370.1 NNAY01000233 OXU29825.1 KQ971330 EFA00972.1 GGFK01008041 MBW41362.1 APGK01049756 KB741156 ENN73619.1 ATLV01024459 KE525352 KFB51423.1 GGFJ01006482 MBW55623.1 GGFM01002794 MBW23545.1 ADMH02001836 ETN60977.1 GFDL01010081 JAV24964.1 GGFJ01006481 MBW55622.1 DS231978 EDS30138.1 KB631941 ERL87376.1 AAAB01008960 GAPW01001984 JAC11614.1 APCN01002286 JXUM01057598 KQ561962 KXJ77051.1 CM000159 EDW94756.2 CH940647 EDW69746.1 AB242867 AE014296 AY069640 CH477309 EAT44004.1 AXCN02000463 CM000363 CM002912 EDX10816.1 KMZ00187.1 JXJN01009712 CH954178 EDV51998.2 AXCM01003551 CH480815 EDW41847.1 CH479211 EDW33093.1 CH379070 KRT08497.1 GECU01009367 JAS98339.1 GAKP01020367 JAC38585.1 CH902618 EDV39585.1 OUUW01000009 SPP85219.1 CH964101 EDW79409.1 LJIJ01000285 ODM99368.1 GDHF01012065 JAI40249.1 GAMC01007118 JAB99437.1 CH916366 EDV96802.1 AMQN01008493 KB303236 ELU03367.1 GECL01000832 JAP05292.1 JRES01000254 KNC32956.1 GBXI01009696 JAD04596.1 GBGD01002086 JAC86803.1 CH933809 EDW18337.2 GEBQ01026053 JAT13924.1 NEVH01016301 PNF25893.1 GGMR01012889 MBY25508.1 LRGB01002722 KZS06460.1 KK854954 PTY19914.1 GDIQ01008627 JAN86110.1 GL732595 EFX72822.1 ACPB03001903 KK852729 KDR17582.1 GFXV01004135 MBW15940.1 GDIP01034394 JAM69321.1 HACG01025774 CEK72639.1 KL894251 KGL81647.1 AHAT01002330 KL218247 KFP03433.1 AABR07027364 AK076592 AK077208 AK141280 BC006881 GDHC01009079 JAQ09550.1 KK827143 KFP73587.1 GBHO01008400 GBHO01001380 GBHO01001379 JAG35204.1 JAG42224.1 JAG42225.1
Proteomes
UP000218220
UP000283053
UP000005204
UP000007151
UP000037510
UP000053240
+ More
UP000053268 UP000215335 UP000007266 UP000002358 UP000019118 UP000030765 UP000000673 UP000075884 UP000002320 UP000030742 UP000075880 UP000007062 UP000075903 UP000076407 UP000075840 UP000069940 UP000249989 UP000002282 UP000075902 UP000008792 UP000000803 UP000192221 UP000008820 UP000075886 UP000000304 UP000075900 UP000092460 UP000008711 UP000075883 UP000001292 UP000076408 UP000008744 UP000001819 UP000075920 UP000007801 UP000075901 UP000075881 UP000268350 UP000007798 UP000094527 UP000075885 UP000001070 UP000014760 UP000037069 UP000095301 UP000095300 UP000009192 UP000235965 UP000076858 UP000076420 UP000000305 UP000015103 UP000027135 UP000078200 UP000092445 UP000085678 UP000053641 UP000265120 UP000018468 UP000005226 UP000054308 UP000002494 UP000000589 UP000221080
UP000053268 UP000215335 UP000007266 UP000002358 UP000019118 UP000030765 UP000000673 UP000075884 UP000002320 UP000030742 UP000075880 UP000007062 UP000075903 UP000076407 UP000075840 UP000069940 UP000249989 UP000002282 UP000075902 UP000008792 UP000000803 UP000192221 UP000008820 UP000075886 UP000000304 UP000075900 UP000092460 UP000008711 UP000075883 UP000001292 UP000076408 UP000008744 UP000001819 UP000075920 UP000007801 UP000075901 UP000075881 UP000268350 UP000007798 UP000094527 UP000075885 UP000001070 UP000014760 UP000037069 UP000095301 UP000095300 UP000009192 UP000235965 UP000076858 UP000076420 UP000000305 UP000015103 UP000027135 UP000078200 UP000092445 UP000085678 UP000053641 UP000265120 UP000018468 UP000005226 UP000054308 UP000002494 UP000000589 UP000221080
Pfam
PF08449 UAA
Interpro
IPR013657
UAA
ProteinModelPortal
D2XRA6
Q2F658
A0A2A4JWN3
A0A3S2NGP5
H9JR97
A0A1E1WA90
+ More
A0A212ERX1 A0A0L7L8N2 A0A194R3S0 A0A194PKX3 A0A0K8TQS0 A0A1Y1LDF0 A0A232FGK7 D6WH53 A0A2M4AKV7 K7J9A9 N6T0M2 A0A084WMH9 A0A2M4BRL8 A0A2M3Z4X2 W5J947 A0A1Q3FBK9 A0A2M4BRH8 A0A182NIY5 B0WKZ8 U4U9X4 A0A182ILV1 Q7Q5D4 A0A182URW5 A0A023ETA5 A0A182X956 A0A182I493 A0A182GE83 B4PJY7 A0A182TGT5 B4LBF8 Q9VVD9 A0A1W4VSK1 Q17CE7 A0A182QTK9 B4QNE6 A0A182RR65 A0A1B0B7S8 B3NDD6 A0A182MU49 B4HKA6 A0A182Y7V4 B4H5P1 A0A0R3P5S5 Q29EY2 A0A182WBY4 A0A1B6JGM3 A0A034V9Y8 B3M5H5 A0A182SJS2 A0A182K706 A0A3B0KFW5 B4N4X0 A0A1D2N287 A0A0K8VMZ2 W8BKZ8 A0A182NZT2 B4J0G6 R7UJB8 A0A0V0GBN0 A0A0L0CL30 A0A1I8MAJ6 A0A0A1X117 A0A1I8P623 A0A069DRV6 B4KV26 A0A1B6KRQ8 A0A2J7QBC8 A0A2S2P8I9 A0A164P2R6 A0A2R7WJU4 A0A0P6I6Q3 A0A2C9LTE5 E9H620 T1HPW5 A0A067R5L8 A0A2H8TPM1 A0A1A9VMZ9 A0A0P6AHT6 A0A1B0A2J2 A0A0B6ZVJ6 A0A1S3H0C6 A0A099ZIK7 A0A3P8VQG3 W5N6L6 A0A3B5KLF8 A0A1S3J8P5 A0A091I7L1 D4AAG3 Q922Q5 A0A146LSY8 A0A091MGF2 A0A2D0PQQ4 A0A0A9YSM8
A0A212ERX1 A0A0L7L8N2 A0A194R3S0 A0A194PKX3 A0A0K8TQS0 A0A1Y1LDF0 A0A232FGK7 D6WH53 A0A2M4AKV7 K7J9A9 N6T0M2 A0A084WMH9 A0A2M4BRL8 A0A2M3Z4X2 W5J947 A0A1Q3FBK9 A0A2M4BRH8 A0A182NIY5 B0WKZ8 U4U9X4 A0A182ILV1 Q7Q5D4 A0A182URW5 A0A023ETA5 A0A182X956 A0A182I493 A0A182GE83 B4PJY7 A0A182TGT5 B4LBF8 Q9VVD9 A0A1W4VSK1 Q17CE7 A0A182QTK9 B4QNE6 A0A182RR65 A0A1B0B7S8 B3NDD6 A0A182MU49 B4HKA6 A0A182Y7V4 B4H5P1 A0A0R3P5S5 Q29EY2 A0A182WBY4 A0A1B6JGM3 A0A034V9Y8 B3M5H5 A0A182SJS2 A0A182K706 A0A3B0KFW5 B4N4X0 A0A1D2N287 A0A0K8VMZ2 W8BKZ8 A0A182NZT2 B4J0G6 R7UJB8 A0A0V0GBN0 A0A0L0CL30 A0A1I8MAJ6 A0A0A1X117 A0A1I8P623 A0A069DRV6 B4KV26 A0A1B6KRQ8 A0A2J7QBC8 A0A2S2P8I9 A0A164P2R6 A0A2R7WJU4 A0A0P6I6Q3 A0A2C9LTE5 E9H620 T1HPW5 A0A067R5L8 A0A2H8TPM1 A0A1A9VMZ9 A0A0P6AHT6 A0A1B0A2J2 A0A0B6ZVJ6 A0A1S3H0C6 A0A099ZIK7 A0A3P8VQG3 W5N6L6 A0A3B5KLF8 A0A1S3J8P5 A0A091I7L1 D4AAG3 Q922Q5 A0A146LSY8 A0A091MGF2 A0A2D0PQQ4 A0A0A9YSM8
Ontologies
GO
Topology
Subcellular location
Golgi apparatus membrane
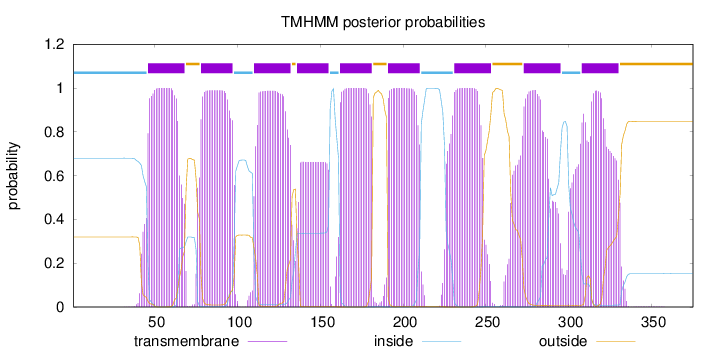
Length:
375
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
190.09027
Exp number, first 60 AAs:
16.13909
Total prob of N-in:
0.67991
POSSIBLE N-term signal
sequence
inside
1 - 45
TMhelix
46 - 68
outside
69 - 77
TMhelix
78 - 97
inside
98 - 109
TMhelix
110 - 132
outside
133 - 135
TMhelix
136 - 155
inside
156 - 161
TMhelix
162 - 181
outside
182 - 190
TMhelix
191 - 210
inside
211 - 230
TMhelix
231 - 253
outside
254 - 272
TMhelix
273 - 295
inside
296 - 307
TMhelix
308 - 330
outside
331 - 375
Population Genetic Test Statistics
Pi
30.585376
Theta
27.254976
Tajima's D
0.414789
CLR
0
CSRT
0.500374981250937
Interpretation
Uncertain
