Gene
KWMTBOMO06831
Pre Gene Modal
BGIBMGA012095
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_MARCH6_isoform_X2_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.679
Sequence
CDS
ATGATGGACTCGGACTCGTCCGGCGCCGACTTCTGCCGGTTTTGTCGCTCGGAGGCAACGTCGGATCGGCCTTTATTCCACCCTTGCATCTGTACCGGTTCGATTAAATGGATCCATCAGGAATGCTTAGTGCAATGGATGAGATACTCTCGGAAGGAAATCTGCGAGCTTTGCGGACATCGCTTTTCTTTTATGCCGATATATTCTCCGGACATGCCTCGCAGGCTGCCTATCCGCGATGTAGTGGGCGGCCTGGTGATGTCCGTTGCGTCAGCCGTAAAGGACTGGCTCCATTACACCCTGGTGGCTTTAGCCTGGCTCGGTATCGTACCGCTTACGGCTTGCAGGACATATAGGTGCTTGTTCTCAGGCTCCCTGGGACCAGTTATTTCCTTACCATTTGAGATTGTTTCTGCTGAGAATCTTGCCAAAGATGTTTTCTCAGGATGTTTTATTGTGACATGTACACTTTTCTCTTTTATTGGACTTGTGTGGTTGAGAGAACAAATAATGCACGGTGGAGGTCCAGACTGGATGGAAAGAGAAAATATACCAGTTCCCCCACCAGAGGATTTTGTACAGCCTGAGAATAATAATGAAAGAGTTAATGAAGTTGGAGGGGATGACGGTGGAGCTCGTGAAGAAGGTAATGGAGTTGGTAACGGTAATGGTGAAGGAGGTGGTGAAGAACCATTAGTGGGCGATGAAGCGAATTGGAATCCAATGGAATGGGATAGAGCTGCTGCTGAAGAATTGACATGGGCCCGACTATTAGGTCTTGATGGGTCCATGGTCTTTCTAGAGCATGTGTTTTGGGTGGTATCCCTCAACACTCTGTTTATAGTGGTGTTTGCATTCTGTCCTTATCACATAGGACGACTTGGGGCTGCCTTAGCAGGATTAGCAACAGAAGGTCCTTATGCTGGACCTTTGACAGCACTGGCTGGCTATGTGTTAGTTGGTGCAATACTTGCCGTACTTCATGGCTTAGCTGCTCTGTTTCGGCTAAGGAGTGCCAAGAAAGCACTAGGATTCTGCTACGTTGTAGTCAAAGTTGCACTGCTTTCAGTCATGGAGATAGGTGTGATCCCTCTAATATGTGGATGGTGGTTGGATCTCTGTTCTCTCTCCATGTTTGATGCGACACTCAAAGACCGCGAATCAAGCCTTCAGGCGGCTCCCTGGACATTGATGTTTATACATTGGCTTGTCGGAATGGTCTATGTGTACTACTTTGCGTCATTTGTGCTGTTACTAAGGGAAGTGCTGAGACCGGGAGTGTTATGGTTCTTGAAGAATCTGAACGATCCTGATTTCAGTCCAGTGCAGGAAATGATTCACTTGTCTGTCATGTGTCACATCAGAAGGCTGGTGTTGTCAGCCATGATCTTTGGCTCGGCAGTACTATTCATGCTTTGGATGCCAATAAGGGTCATTAAATATGTGCTGCCCGGTTTCCTGCCATACACTGTCTCCGTTCATTCAGAAGCCCCAGTCAATGAATTAAGTTTGGAACTTCTTCTGCTTCAGGTGATCCTTCCAGCTCTTCTGGAGCAATCTCACACGAGGACGTGGTTAAAGATGGCGGTGCGAGCTTGGTGCGTGTGCGCTTCTGGTCTCCTCGGCTTGAGATCGTATCTGCTTGGCGAGATATCGCGACAGGCCGATCCTAATCCGCCCCCACATCTGCCTCATCAACTCGGTGTCGCTCATCAAGCCCTGATGCGTCGCGACGGTCCAGCGGCGGTGGAGCCCTACATCCGCGTGTCGTGGTTCCCGCTAAGGTTGGCCGCGCTGCTTGTGCTGGTCTCTATTTCGCTCGTGTTAGCGAGTGCAATGACCCTTGTAATTCCAGTAGCGATCGGCAGAAAGGTGATGGCGCTGTGGATGCCTAAGGCGGCTGAGAGAGTTCACGAACTGTATACCGCTGCATGCGGTATGTACGTGAGCTGGGCGGCGGGGCGGGCGGGCGTGGCGGCGGCGGGGTGGGCGCGGGGCGGGGCGCGCGGGGGCCTGTGGTCGCGCGCCGCGGTGTGGGCCCGCACGGCCGCCCGCGCCGCGCTCGCCGCCGTGCTACTGCTCGGGCTGGTGCCGCTCATGTTCGGCTTGCTGCTCGAACTGGTTCTCGTAATACCACTGAGCGTTCCCTTGGAGCAGTCCCCTGTGTTATTCGTGTGGCAAGATTGGGCTCTGGGCGTGCTGTATACGAAGATAATGTGCGCCCTCACGATGATGGGACCCGATTGGAGCATGCGCCGTGCCCTCGAGAAAGCGTACCGGGACGGCATCAGAGAAATGGATTTAAGGTTTATCGTGGGGTCGGTGGGTGCCCCGCTGGTGCGGTGGCTGGGGCTGGCGCTGGCGGTGCCGTACGCGGCGGCGCACAGCGTGGCGCCCGTGCTGGTGTCAGGGGCCGCCCAGCGCAACCTGCTCGTGCGCCGCCTCTACCCCGCGCTGCTCCTCGCCGCGCTCTTCGCCTCGCTAGCGGCCTTCCAGATCCGCCAATTCAGAAAACTATACGAACACATAAAGAACGACAAGTACCTGGTAGGTCAGCGGCTCGTGAACTACGATCACCGGAGACAGAAGCAGCAGCACAGCAGTGCCGTCACGTCGACAAACTGA
Protein
MMDSDSSGADFCRFCRSEATSDRPLFHPCICTGSIKWIHQECLVQWMRYSRKEICELCGHRFSFMPIYSPDMPRRLPIRDVVGGLVMSVASAVKDWLHYTLVALAWLGIVPLTACRTYRCLFSGSLGPVISLPFEIVSAENLAKDVFSGCFIVTCTLFSFIGLVWLREQIMHGGGPDWMERENIPVPPPEDFVQPENNNERVNEVGGDDGGAREEGNGVGNGNGEGGGEEPLVGDEANWNPMEWDRAAAEELTWARLLGLDGSMVFLEHVFWVVSLNTLFIVVFAFCPYHIGRLGAALAGLATEGPYAGPLTALAGYVLVGAILAVLHGLAALFRLRSAKKALGFCYVVVKVALLSVMEIGVIPLICGWWLDLCSLSMFDATLKDRESSLQAAPWTLMFIHWLVGMVYVYYFASFVLLLREVLRPGVLWFLKNLNDPDFSPVQEMIHLSVMCHIRRLVLSAMIFGSAVLFMLWMPIRVIKYVLPGFLPYTVSVHSEAPVNELSLELLLLQVILPALLEQSHTRTWLKMAVRAWCVCASGLLGLRSYLLGEISRQADPNPPPHLPHQLGVAHQALMRRDGPAAVEPYIRVSWFPLRLAALLVLVSISLVLASAMTLVIPVAIGRKVMALWMPKAAERVHELYTAACGMYVSWAAGRAGVAAAGWARGGARGGLWSRAAVWARTAARAALAAVLLLGLVPLMFGLLLELVLVIPLSVPLEQSPVLFVWQDWALGVLYTKIMCALTMMGPDWSMRRALEKAYRDGIREMDLRFIVGSVGAPLVRWLGLALAVPYAAAHSVAPVLVSGAAQRNLLVRRLYPALLLAALFASLAAFQIRQFRKLYEHIKNDKYLVGQRLVNYDHRRQKQQHSSAVTSTN
Summary
Uniprot
A0A2H1W1K3
A0A194PGC4
A0A1W4WTW8
A0A1Y1MW80
A0A2P8ZKG7
G3UN33
+ More
D6WXQ6 A0A1B6ML90 A0A1B6CFS0 A0A310STL8 A0A0T6B8Y6 R4WPS5 A0A067R7K2 U3J0M2 W5N7T3 A0A3P8UA77 A0A093IEL8 A0A3P8YB45 A0A1S6K828 A0A1S3R268 A0A1S3R0R7 A0A1S3R0Q9 A0A1S3R1D6 A0A3P8YB81 A0A0B8RPL8 A0A1S3LYE8 A2RV74 A0A1S3LY71 A0A091HV00 A0A1L8FX62 M3YIH9 E9QDQ9 V9KDE1 A0A218UHC0 A0A3Q2YY25 A0A1W5A8W8 A0A3Q1IJ82 A0A3P9JM15 A0A091FKF4 G3SZL8 A0A3B3WZJ3 A0A087X849 A0A3P9PGU5 U3JZD4 A0A3Q0G3R3 A0A0S7FQI1 M4AXL0 A0A3Q0DAH2 A0A091QF32 A0A3B3TSQ5 A0A3B3XBN2 A0A1V4JWG8 G1N311 A0A1V4JW97 A0A2U4CMH1 A0A2Y9SC72 A0A0M9A862 A0A091P8Z2 A0A2Y9NVW6 A0A091SUB5 A0A099ZB72 A0A091WLJ7 A0A3Q2U218 A0A2U4CMG3 A0A340X8Y5 H3CLH0 A0A093H9F0 A0A0F7Z3C3 A0A2D4PHY6 A0A2I0URH3 A0A3P9L7E1 A0A2D4FEU4 T1E481 A0A3P9MX19 A0A2I0MWC4 A0A2U4CMG7 A0A091L127 A0A341D7V3 A0A2U4CMG9 A0A2Y9NSR7 A0A3Q3J8Y7 A0A091FP21 A0A1D5PSM7 A0A091J872 A0A091T535 A0A093FSL4 A0A087VF95 A0A1D5NWV0 A0A3Q3QI95 A0A093P679 A0A1A8UDC9 A0A1A8P458 A0A1A8MXJ9 A0A1A8I6K7 A0A1A8DPW2 A0A1A7X590
D6WXQ6 A0A1B6ML90 A0A1B6CFS0 A0A310STL8 A0A0T6B8Y6 R4WPS5 A0A067R7K2 U3J0M2 W5N7T3 A0A3P8UA77 A0A093IEL8 A0A3P8YB45 A0A1S6K828 A0A1S3R268 A0A1S3R0R7 A0A1S3R0Q9 A0A1S3R1D6 A0A3P8YB81 A0A0B8RPL8 A0A1S3LYE8 A2RV74 A0A1S3LY71 A0A091HV00 A0A1L8FX62 M3YIH9 E9QDQ9 V9KDE1 A0A218UHC0 A0A3Q2YY25 A0A1W5A8W8 A0A3Q1IJ82 A0A3P9JM15 A0A091FKF4 G3SZL8 A0A3B3WZJ3 A0A087X849 A0A3P9PGU5 U3JZD4 A0A3Q0G3R3 A0A0S7FQI1 M4AXL0 A0A3Q0DAH2 A0A091QF32 A0A3B3TSQ5 A0A3B3XBN2 A0A1V4JWG8 G1N311 A0A1V4JW97 A0A2U4CMH1 A0A2Y9SC72 A0A0M9A862 A0A091P8Z2 A0A2Y9NVW6 A0A091SUB5 A0A099ZB72 A0A091WLJ7 A0A3Q2U218 A0A2U4CMG3 A0A340X8Y5 H3CLH0 A0A093H9F0 A0A0F7Z3C3 A0A2D4PHY6 A0A2I0URH3 A0A3P9L7E1 A0A2D4FEU4 T1E481 A0A3P9MX19 A0A2I0MWC4 A0A2U4CMG7 A0A091L127 A0A341D7V3 A0A2U4CMG9 A0A2Y9NSR7 A0A3Q3J8Y7 A0A091FP21 A0A1D5PSM7 A0A091J872 A0A091T535 A0A093FSL4 A0A087VF95 A0A1D5NWV0 A0A3Q3QI95 A0A093P679 A0A1A8UDC9 A0A1A8P458 A0A1A8MXJ9 A0A1A8I6K7 A0A1A8DPW2 A0A1A7X590
Pubmed
EMBL
ODYU01005485
SOQ46384.1
KQ459606
KPI91754.1
GEZM01021923
JAV88820.1
+ More
PYGN01000029 PSN56991.1 KQ971362 EFA08890.2 GEBQ01003315 JAT36662.1 GEDC01025113 GEDC01002090 JAS12185.1 JAS35208.1 KQ760382 OAD60712.1 LJIG01009089 KRT83778.1 AK417671 BAN20886.1 KK852649 KDR19487.1 ADON01115497 AHAT01033574 AHAT01033575 KK590893 KFW00038.1 KX531015 AQS99344.1 GBSH01003012 JAG66015.1 BC133209 AAI33210.1 KL217809 KFO98989.1 CM004476 OCT76173.1 AEYP01051773 AEYP01051774 AEYP01051775 CR388419 JW863290 AFO95807.1 MUZQ01000301 OWK53187.1 KK719183 KFO61830.1 AYCK01002802 AGTO01021551 GBYX01462283 JAO19302.1 KK695776 KFQ24221.1 LSYS01005643 OPJ76481.1 OPJ76482.1 KQ435714 KOX79197.1 KK654719 KFQ04080.1 KK486873 KFQ62254.1 KL891024 KGL78278.1 KK735815 KFR15995.1 AADN05000026 KL205871 KFV75647.1 GBEX01003832 JAI10728.1 IACN01072482 LAB57088.1 KZ505649 PKU48638.1 IACJ01065132 LAA46005.1 GAAZ01002816 GBKC01002018 JAA95127.1 JAG44052.1 AKCR02000001 PKK33971.1 KL290946 KFP49527.1 KL447194 KFO70726.1 KK501545 KFP15920.1 KK438479 KFQ69486.1 KK633841 KFV57319.1 KL491101 KFO11287.1 KL225137 KFW67752.1 HADY01005820 HAEJ01005847 SBS46304.1 HAEH01005269 HAEI01010432 SBR76076.1 HAEF01020446 HAEG01000788 SBR61605.1 HAED01006227 HAEE01002401 SBQ92257.1 HADZ01013182 HAEA01006379 SBQ34859.1 HADW01011718 HADX01015250 SBP13118.1
PYGN01000029 PSN56991.1 KQ971362 EFA08890.2 GEBQ01003315 JAT36662.1 GEDC01025113 GEDC01002090 JAS12185.1 JAS35208.1 KQ760382 OAD60712.1 LJIG01009089 KRT83778.1 AK417671 BAN20886.1 KK852649 KDR19487.1 ADON01115497 AHAT01033574 AHAT01033575 KK590893 KFW00038.1 KX531015 AQS99344.1 GBSH01003012 JAG66015.1 BC133209 AAI33210.1 KL217809 KFO98989.1 CM004476 OCT76173.1 AEYP01051773 AEYP01051774 AEYP01051775 CR388419 JW863290 AFO95807.1 MUZQ01000301 OWK53187.1 KK719183 KFO61830.1 AYCK01002802 AGTO01021551 GBYX01462283 JAO19302.1 KK695776 KFQ24221.1 LSYS01005643 OPJ76481.1 OPJ76482.1 KQ435714 KOX79197.1 KK654719 KFQ04080.1 KK486873 KFQ62254.1 KL891024 KGL78278.1 KK735815 KFR15995.1 AADN05000026 KL205871 KFV75647.1 GBEX01003832 JAI10728.1 IACN01072482 LAB57088.1 KZ505649 PKU48638.1 IACJ01065132 LAA46005.1 GAAZ01002816 GBKC01002018 JAA95127.1 JAG44052.1 AKCR02000001 PKK33971.1 KL290946 KFP49527.1 KL447194 KFO70726.1 KK501545 KFP15920.1 KK438479 KFQ69486.1 KK633841 KFV57319.1 KL491101 KFO11287.1 KL225137 KFW67752.1 HADY01005820 HAEJ01005847 SBS46304.1 HAEH01005269 HAEI01010432 SBR76076.1 HAEF01020446 HAEG01000788 SBR61605.1 HAED01006227 HAEE01002401 SBQ92257.1 HADZ01013182 HAEA01006379 SBQ34859.1 HADW01011718 HADX01015250 SBP13118.1
Proteomes
UP000053268
UP000192223
UP000245037
UP000007646
UP000007266
UP000027135
+ More
UP000016666 UP000018468 UP000265080 UP000265140 UP000087266 UP000054308 UP000186698 UP000000715 UP000000437 UP000197619 UP000264820 UP000192224 UP000265040 UP000265200 UP000052976 UP000261480 UP000028760 UP000242638 UP000016665 UP000189705 UP000002852 UP000189706 UP000261500 UP000190648 UP000001645 UP000245320 UP000248484 UP000053105 UP000248483 UP000053641 UP000053605 UP000000539 UP000265300 UP000007303 UP000053584 UP000265180 UP000053872 UP000252040 UP000261600 UP000053760 UP000053119 UP000054081
UP000016666 UP000018468 UP000265080 UP000265140 UP000087266 UP000054308 UP000186698 UP000000715 UP000000437 UP000197619 UP000264820 UP000192224 UP000265040 UP000265200 UP000052976 UP000261480 UP000028760 UP000242638 UP000016665 UP000189705 UP000002852 UP000189706 UP000261500 UP000190648 UP000001645 UP000245320 UP000248484 UP000053105 UP000248483 UP000053641 UP000053605 UP000000539 UP000265300 UP000007303 UP000053584 UP000265180 UP000053872 UP000252040 UP000261600 UP000053760 UP000053119 UP000054081
Pfam
PF12906 RINGv
Gene 3D
ProteinModelPortal
A0A2H1W1K3
A0A194PGC4
A0A1W4WTW8
A0A1Y1MW80
A0A2P8ZKG7
G3UN33
+ More
D6WXQ6 A0A1B6ML90 A0A1B6CFS0 A0A310STL8 A0A0T6B8Y6 R4WPS5 A0A067R7K2 U3J0M2 W5N7T3 A0A3P8UA77 A0A093IEL8 A0A3P8YB45 A0A1S6K828 A0A1S3R268 A0A1S3R0R7 A0A1S3R0Q9 A0A1S3R1D6 A0A3P8YB81 A0A0B8RPL8 A0A1S3LYE8 A2RV74 A0A1S3LY71 A0A091HV00 A0A1L8FX62 M3YIH9 E9QDQ9 V9KDE1 A0A218UHC0 A0A3Q2YY25 A0A1W5A8W8 A0A3Q1IJ82 A0A3P9JM15 A0A091FKF4 G3SZL8 A0A3B3WZJ3 A0A087X849 A0A3P9PGU5 U3JZD4 A0A3Q0G3R3 A0A0S7FQI1 M4AXL0 A0A3Q0DAH2 A0A091QF32 A0A3B3TSQ5 A0A3B3XBN2 A0A1V4JWG8 G1N311 A0A1V4JW97 A0A2U4CMH1 A0A2Y9SC72 A0A0M9A862 A0A091P8Z2 A0A2Y9NVW6 A0A091SUB5 A0A099ZB72 A0A091WLJ7 A0A3Q2U218 A0A2U4CMG3 A0A340X8Y5 H3CLH0 A0A093H9F0 A0A0F7Z3C3 A0A2D4PHY6 A0A2I0URH3 A0A3P9L7E1 A0A2D4FEU4 T1E481 A0A3P9MX19 A0A2I0MWC4 A0A2U4CMG7 A0A091L127 A0A341D7V3 A0A2U4CMG9 A0A2Y9NSR7 A0A3Q3J8Y7 A0A091FP21 A0A1D5PSM7 A0A091J872 A0A091T535 A0A093FSL4 A0A087VF95 A0A1D5NWV0 A0A3Q3QI95 A0A093P679 A0A1A8UDC9 A0A1A8P458 A0A1A8MXJ9 A0A1A8I6K7 A0A1A8DPW2 A0A1A7X590
D6WXQ6 A0A1B6ML90 A0A1B6CFS0 A0A310STL8 A0A0T6B8Y6 R4WPS5 A0A067R7K2 U3J0M2 W5N7T3 A0A3P8UA77 A0A093IEL8 A0A3P8YB45 A0A1S6K828 A0A1S3R268 A0A1S3R0R7 A0A1S3R0Q9 A0A1S3R1D6 A0A3P8YB81 A0A0B8RPL8 A0A1S3LYE8 A2RV74 A0A1S3LY71 A0A091HV00 A0A1L8FX62 M3YIH9 E9QDQ9 V9KDE1 A0A218UHC0 A0A3Q2YY25 A0A1W5A8W8 A0A3Q1IJ82 A0A3P9JM15 A0A091FKF4 G3SZL8 A0A3B3WZJ3 A0A087X849 A0A3P9PGU5 U3JZD4 A0A3Q0G3R3 A0A0S7FQI1 M4AXL0 A0A3Q0DAH2 A0A091QF32 A0A3B3TSQ5 A0A3B3XBN2 A0A1V4JWG8 G1N311 A0A1V4JW97 A0A2U4CMH1 A0A2Y9SC72 A0A0M9A862 A0A091P8Z2 A0A2Y9NVW6 A0A091SUB5 A0A099ZB72 A0A091WLJ7 A0A3Q2U218 A0A2U4CMG3 A0A340X8Y5 H3CLH0 A0A093H9F0 A0A0F7Z3C3 A0A2D4PHY6 A0A2I0URH3 A0A3P9L7E1 A0A2D4FEU4 T1E481 A0A3P9MX19 A0A2I0MWC4 A0A2U4CMG7 A0A091L127 A0A341D7V3 A0A2U4CMG9 A0A2Y9NSR7 A0A3Q3J8Y7 A0A091FP21 A0A1D5PSM7 A0A091J872 A0A091T535 A0A093FSL4 A0A087VF95 A0A1D5NWV0 A0A3Q3QI95 A0A093P679 A0A1A8UDC9 A0A1A8P458 A0A1A8MXJ9 A0A1A8I6K7 A0A1A8DPW2 A0A1A7X590
PDB
2M6M
E-value=1.01527e-11,
Score=172
Ontologies
GO
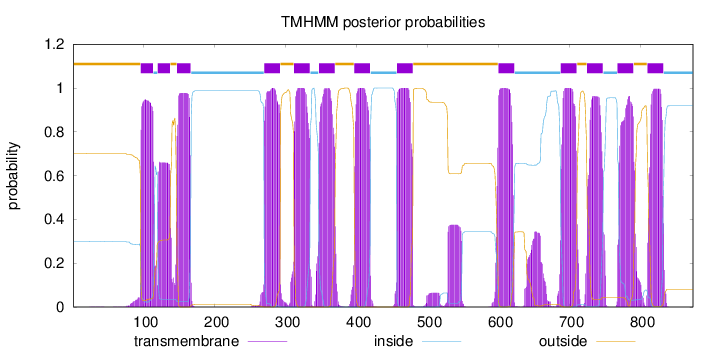
Topology
Length:
874
Number of predicted TMHs:
13
Exp number of AAs in TMHs:
291.69216
Exp number, first 60 AAs:
0.00804
Total prob of N-in:
0.29861
outside
1 - 95
TMhelix
96 - 113
inside
114 - 119
TMhelix
120 - 137
outside
138 - 146
TMhelix
147 - 166
inside
167 - 269
TMhelix
270 - 292
outside
293 - 311
TMhelix
312 - 334
inside
335 - 346
TMhelix
347 - 369
outside
370 - 396
TMhelix
397 - 419
inside
420 - 456
TMhelix
457 - 479
outside
480 - 599
TMhelix
600 - 622
inside
623 - 687
TMhelix
688 - 710
outside
711 - 724
TMhelix
725 - 747
inside
748 - 767
TMhelix
768 - 790
outside
791 - 809
TMhelix
810 - 832
inside
833 - 874
Population Genetic Test Statistics
Pi
26.4183
Theta
21.810205
Tajima's D
-1.40852
CLR
0.022163
CSRT
0.0706464676766162
Interpretation
Uncertain
