Gene
KWMTBOMO06827
Pre Gene Modal
BGIBMGA012054
Annotation
PREDICTED:_protein_N-terminal_asparagine_amidohydrolase_isoform_X1_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.25 PlasmaMembrane Reliability : 1.018
Sequence
CDS
ATGAAGACGAACTCGCGTTGCGCGGGTGCCAGGATCCTAATCGCTTACAAACATTCCAGCGGCATCATGGTGGTGGTGCTGAACGGCGTACTCGCTGACGATTGCCCGACGTCGGTGCGGGCTTTGCTAGAGGCGCATCCGGGGTACAGAGAAGGCGCCGCGCAACTGTTGACCGAAACGGCGCGCGTTGTGGGACCCGCGGGCGTTCTTTACGTCGGACAGAGAGAAATGGCCGCCGTTGTGCCGCATGACAAAAACGTGAATATAATCGGTTCAGATGACGCGACATCGTGTATTATAGTAGTCGTCAGGCATTCAGGGTCAGGGGCCATAGCGCTAGCGCACTTGGACGGTTCGGGTACGGCTGAAGCGGCAGCGGCCATGGTGTCCAGAGTCCAGCAGCTCGCAGTCGGATATCCTGAGGGTAGACTGGAGTTGCAAGTGATCGGCGGCTTCACGGATCCACATAGATACTCCGACGAGCTTTTCGCTAATATTATGTTATCATTCCATCGTCTAACGGTAGAGATAGACCTGACGTTGGCGTGTTGCTGCGAGCTGAACACGCTGCCCGGGGGACTGGCGCCTCTCGTGACCGGGGTCGGCGTCGACATCCGCGCCGGGGATCTCTTCCCGGCAACGTTTCCGGATAAAGGTCCCGAACTTCCGTTGCGAGGGGCGAGAACTATAACGAGCGGTCCGCATTCGGCACAGGTGTTAGACATATACGATAACGCTGTGGGTATGCTACGCGTCGGCCCGTTCAACTACGACCCCCTGCGAGGAGTCGACCTCTGGCTCGAACAGTCTGATGAGTTCATTCTGCAGCACCTAAGCACGACGCCGGCCGTCGAGCCGCCGCATTTCGTTCATAATATCCGGATGACGCTGAAGTTCATCCAGCAGCATCCGTTTCCGGCGGTGACCGTGTTCCCGGATAACCGGCCGCACTTCTACCGCCGCGACGAGCACTCCGGTTGTTGGATATCCGTTCGTTACTAA
Protein
MKTNSRCAGARILIAYKHSSGIMVVVLNGVLADDCPTSVRALLEAHPGYREGAAQLLTETARVVGPAGVLYVGQREMAAVVPHDKNVNIIGSDDATSCIIVVVRHSGSGAIALAHLDGSGTAEAAAAMVSRVQQLAVGYPEGRLELQVIGGFTDPHRYSDELFANIMLSFHRLTVEIDLTLACCCELNTLPGGLAPLVTGVGVDIRAGDLFPATFPDKGPELPLRGARTITSGPHSAQVLDIYDNAVGMLRVGPFNYDPLRGVDLWLEQSDEFILQHLSTTPAVEPPHFVHNIRMTLKFIQQHPFPAVTVFPDNRPHFYRRDEHSGCWISVRY
Summary
Uniprot
H9JR94
A0A2A4J421
A0A2H1VA15
A0A212F420
A0A437BJK9
A0A194PEQ9
+ More
A0A194R3S5 D6WDI5 A0A2J7RQ33 A0A1L8E2M8 U5ESL8 A0A1Y1LRQ0 A0A084WQY8 A0A1Q3FH91 A0A182QEE0 A0A182NA40 A0A182PJG6 A0A182R2Y2 A0A182KKD7 A0A182WFU7 A0A182WUM1 A0A182V0C7 B0XAF5 A0A182H4E5 Q17BH8 A0A023EQG7 A0A182JJI4 A0A1Q3FHQ3 A0A2M4CV54 A0A2M4ASY3 A0A2P8Y3E9 A0A182FKE9 A0A2M3Z9R4 A0A2A3E1N8 A0A0L7R0I2 A0A088AP41 A0A0M9A056 E2BEF3 A0A0A1XAY0 K7IPK1 A0A0C9R7S0 A0A232FBE9 T1PKI9 A0A067R6F7 W8C2F4 N6U2W7 J3JUB2 A0A0K8TXW5 A0A034W0Q6 A0A151JVW1 A0A1I8NC15 A0A151WZJ2 E9INW6 F4WH33 A0A195BT04 A0A158NT88 A0A195E355 B4J7N7 A0A154PQR0 B4MRB8 D3TMG3 A0A1A9V8M2 A0A1B6JR27 B4LJ59 B4KPM2 A0A0K8V5E4 A0A034VZ41 A0A1B0G2F3 A0A1I8Q0I7 A0A1I8Q0L4 A0A1A9X2Z3 A0A1B6LLI3 A0A1B6IDW3 A0A0L0CRV6 A0A0P8ZMP3 A0A1W4UM73 A0A1A9ZVI7 A0A0Q9WPI6 A0A3B0J2U3 B4GA64 A0A0R3NLH0 A0A0M4EAG3 A0A0R1DRY6 A0A1B6CM09 A0A0J9RG25 A0A0Q5VKU4 Q7KIS4 E2A596 A0A1W4UMD8 B3MBU5 Q291W5 A0A1A9XAE8 B4P4B3 T1HP56 B4HNZ4 B4QD07 B3NK92 Q8MSD1 A0A026X1Q3 A0A224XVA2
A0A194R3S5 D6WDI5 A0A2J7RQ33 A0A1L8E2M8 U5ESL8 A0A1Y1LRQ0 A0A084WQY8 A0A1Q3FH91 A0A182QEE0 A0A182NA40 A0A182PJG6 A0A182R2Y2 A0A182KKD7 A0A182WFU7 A0A182WUM1 A0A182V0C7 B0XAF5 A0A182H4E5 Q17BH8 A0A023EQG7 A0A182JJI4 A0A1Q3FHQ3 A0A2M4CV54 A0A2M4ASY3 A0A2P8Y3E9 A0A182FKE9 A0A2M3Z9R4 A0A2A3E1N8 A0A0L7R0I2 A0A088AP41 A0A0M9A056 E2BEF3 A0A0A1XAY0 K7IPK1 A0A0C9R7S0 A0A232FBE9 T1PKI9 A0A067R6F7 W8C2F4 N6U2W7 J3JUB2 A0A0K8TXW5 A0A034W0Q6 A0A151JVW1 A0A1I8NC15 A0A151WZJ2 E9INW6 F4WH33 A0A195BT04 A0A158NT88 A0A195E355 B4J7N7 A0A154PQR0 B4MRB8 D3TMG3 A0A1A9V8M2 A0A1B6JR27 B4LJ59 B4KPM2 A0A0K8V5E4 A0A034VZ41 A0A1B0G2F3 A0A1I8Q0I7 A0A1I8Q0L4 A0A1A9X2Z3 A0A1B6LLI3 A0A1B6IDW3 A0A0L0CRV6 A0A0P8ZMP3 A0A1W4UM73 A0A1A9ZVI7 A0A0Q9WPI6 A0A3B0J2U3 B4GA64 A0A0R3NLH0 A0A0M4EAG3 A0A0R1DRY6 A0A1B6CM09 A0A0J9RG25 A0A0Q5VKU4 Q7KIS4 E2A596 A0A1W4UMD8 B3MBU5 Q291W5 A0A1A9XAE8 B4P4B3 T1HP56 B4HNZ4 B4QD07 B3NK92 Q8MSD1 A0A026X1Q3 A0A224XVA2
Pubmed
19121390
22118469
26354079
18362917
19820115
28004739
+ More
24438588 20966253 26483478 17510324 24945155 29403074 20798317 25830018 20075255 28648823 25315136 24845553 24495485 23537049 22516182 25348373 21282665 21719571 21347285 17994087 20353571 18057021 26108605 15632085 23185243 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741
24438588 20966253 26483478 17510324 24945155 29403074 20798317 25830018 20075255 28648823 25315136 24845553 24495485 23537049 22516182 25348373 21282665 21719571 21347285 17994087 20353571 18057021 26108605 15632085 23185243 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741
EMBL
BABH01016254
NWSH01003536
PCG66152.1
ODYU01001196
SOQ37074.1
AGBW02010454
+ More
OWR48479.1 RSAL01000045 RVE50621.1 KQ459606 KPI91762.1 KQ460779 KPJ12342.1 KQ971322 EFA00780.2 NEVH01001347 PNF42957.1 GFDF01001094 JAV12990.1 GANO01002292 JAB57579.1 GEZM01050255 JAV75651.1 ATLV01025844 ATLV01025845 ATLV01025846 ATLV01025847 KE525401 KFB52632.1 GFDL01008140 JAV26905.1 AXCN02002379 DS232581 EDS43652.1 JXUM01109464 JXUM01109465 JXUM01109466 KQ565323 KXJ71088.1 CH477321 EAT43632.1 JXUM01089919 GAPW01002779 KQ563804 JAC10819.1 KXJ73259.1 GFDL01007958 JAV27087.1 GGFL01004530 MBW68708.1 GGFK01010563 MBW43884.1 PYGN01000973 PSN38797.1 GGFM01004469 MBW25220.1 KZ288483 PBC25242.1 KQ414670 KOC64338.1 KQ435783 KOX74650.1 GL447768 EFN85998.1 GBXI01011109 GBXI01005833 JAD03183.1 JAD08459.1 GBYB01008887 GBYB01008888 GBYB01008890 JAG78654.1 JAG78655.1 JAG78657.1 NNAY01000545 OXU27768.1 KA648610 AFP63239.1 KK852855 KDR14937.1 GAMC01010636 GAMC01010635 GAMC01010634 JAB95919.1 APGK01044528 APGK01044529 KB741026 ENN74956.1 BT126825 AEE61787.1 GDHF01033188 GDHF01011509 JAI19126.1 JAI40805.1 GAKP01011594 GAKP01011592 GAKP01011591 GAKP01011589 JAC47358.1 KQ981686 KYN37950.1 KQ982636 KYQ53313.1 GL764397 EFZ17709.1 GL888148 EGI66512.1 KQ976409 KYM90927.1 ADTU01025720 KQ979701 KYN19578.1 CH916367 EDW02185.1 KQ435007 KZC13684.1 CH963850 EDW74657.1 EZ422615 ADD18891.1 GECU01018636 GECU01006010 JAS89070.1 JAT01697.1 CH940648 EDW61495.1 KRF80013.1 CH933808 EDW09132.1 KRG04462.1 KRG04463.1 GDHF01018341 GDHF01000581 JAI33973.1 JAI51733.1 GAKP01011593 GAKP01011590 JAC47362.1 CCAG010006799 GEBQ01021559 GEBQ01017650 GEBQ01015480 JAT18418.1 JAT22327.1 JAT24497.1 GECU01022614 JAS85092.1 JRES01000092 KNC34164.1 CH902619 KPU76012.1 KPU76013.1 KRF98115.1 OUUW01000001 SPP73492.1 CH479181 EDW31816.1 CM000071 KRT01769.1 KRT01770.1 CP012524 ALC42058.1 CM000158 KRJ99982.1 KRJ99983.1 GEDC01022883 GEDC01009118 JAS14415.1 JAS28180.1 CM002911 KMY94907.1 CH954179 KQS62099.1 AF239612 AE013599 AAF63504.1 ACZ94471.1 ACZ94472.1 GL436894 EFN71392.1 EDV36116.2 EAL24997.3 EDW91599.1 KRJ99981.1 ACPB03013520 CH480816 EDW48496.1 CM000362 EDX07712.1 KMY94905.1 EDV55114.1 KQS62098.1 AY118895 AAF57658.2 AAF57659.2 AAM50755.1 KK107061 QOIP01000014 EZA61324.1 RLU14757.1 GFTR01004565 JAW11861.1
OWR48479.1 RSAL01000045 RVE50621.1 KQ459606 KPI91762.1 KQ460779 KPJ12342.1 KQ971322 EFA00780.2 NEVH01001347 PNF42957.1 GFDF01001094 JAV12990.1 GANO01002292 JAB57579.1 GEZM01050255 JAV75651.1 ATLV01025844 ATLV01025845 ATLV01025846 ATLV01025847 KE525401 KFB52632.1 GFDL01008140 JAV26905.1 AXCN02002379 DS232581 EDS43652.1 JXUM01109464 JXUM01109465 JXUM01109466 KQ565323 KXJ71088.1 CH477321 EAT43632.1 JXUM01089919 GAPW01002779 KQ563804 JAC10819.1 KXJ73259.1 GFDL01007958 JAV27087.1 GGFL01004530 MBW68708.1 GGFK01010563 MBW43884.1 PYGN01000973 PSN38797.1 GGFM01004469 MBW25220.1 KZ288483 PBC25242.1 KQ414670 KOC64338.1 KQ435783 KOX74650.1 GL447768 EFN85998.1 GBXI01011109 GBXI01005833 JAD03183.1 JAD08459.1 GBYB01008887 GBYB01008888 GBYB01008890 JAG78654.1 JAG78655.1 JAG78657.1 NNAY01000545 OXU27768.1 KA648610 AFP63239.1 KK852855 KDR14937.1 GAMC01010636 GAMC01010635 GAMC01010634 JAB95919.1 APGK01044528 APGK01044529 KB741026 ENN74956.1 BT126825 AEE61787.1 GDHF01033188 GDHF01011509 JAI19126.1 JAI40805.1 GAKP01011594 GAKP01011592 GAKP01011591 GAKP01011589 JAC47358.1 KQ981686 KYN37950.1 KQ982636 KYQ53313.1 GL764397 EFZ17709.1 GL888148 EGI66512.1 KQ976409 KYM90927.1 ADTU01025720 KQ979701 KYN19578.1 CH916367 EDW02185.1 KQ435007 KZC13684.1 CH963850 EDW74657.1 EZ422615 ADD18891.1 GECU01018636 GECU01006010 JAS89070.1 JAT01697.1 CH940648 EDW61495.1 KRF80013.1 CH933808 EDW09132.1 KRG04462.1 KRG04463.1 GDHF01018341 GDHF01000581 JAI33973.1 JAI51733.1 GAKP01011593 GAKP01011590 JAC47362.1 CCAG010006799 GEBQ01021559 GEBQ01017650 GEBQ01015480 JAT18418.1 JAT22327.1 JAT24497.1 GECU01022614 JAS85092.1 JRES01000092 KNC34164.1 CH902619 KPU76012.1 KPU76013.1 KRF98115.1 OUUW01000001 SPP73492.1 CH479181 EDW31816.1 CM000071 KRT01769.1 KRT01770.1 CP012524 ALC42058.1 CM000158 KRJ99982.1 KRJ99983.1 GEDC01022883 GEDC01009118 JAS14415.1 JAS28180.1 CM002911 KMY94907.1 CH954179 KQS62099.1 AF239612 AE013599 AAF63504.1 ACZ94471.1 ACZ94472.1 GL436894 EFN71392.1 EDV36116.2 EAL24997.3 EDW91599.1 KRJ99981.1 ACPB03013520 CH480816 EDW48496.1 CM000362 EDX07712.1 KMY94905.1 EDV55114.1 KQS62098.1 AY118895 AAF57658.2 AAF57659.2 AAM50755.1 KK107061 QOIP01000014 EZA61324.1 RLU14757.1 GFTR01004565 JAW11861.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000007266 UP000235965 UP000030765 UP000075886 UP000075884 UP000075885 UP000075900 UP000075882 UP000075920 UP000076407 UP000075903 UP000002320 UP000069940 UP000249989 UP000008820 UP000075880 UP000245037 UP000069272 UP000242457 UP000053825 UP000005203 UP000053105 UP000008237 UP000002358 UP000215335 UP000095301 UP000027135 UP000019118 UP000078541 UP000075809 UP000007755 UP000078540 UP000005205 UP000078492 UP000001070 UP000076502 UP000007798 UP000078200 UP000008792 UP000009192 UP000092444 UP000095300 UP000091820 UP000037069 UP000007801 UP000192221 UP000092445 UP000268350 UP000008744 UP000001819 UP000092553 UP000002282 UP000008711 UP000000803 UP000000311 UP000092443 UP000015103 UP000001292 UP000000304 UP000053097 UP000279307
UP000007266 UP000235965 UP000030765 UP000075886 UP000075884 UP000075885 UP000075900 UP000075882 UP000075920 UP000076407 UP000075903 UP000002320 UP000069940 UP000249989 UP000008820 UP000075880 UP000245037 UP000069272 UP000242457 UP000053825 UP000005203 UP000053105 UP000008237 UP000002358 UP000215335 UP000095301 UP000027135 UP000019118 UP000078541 UP000075809 UP000007755 UP000078540 UP000005205 UP000078492 UP000001070 UP000076502 UP000007798 UP000078200 UP000008792 UP000009192 UP000092444 UP000095300 UP000091820 UP000037069 UP000007801 UP000192221 UP000092445 UP000268350 UP000008744 UP000001819 UP000092553 UP000002282 UP000008711 UP000000803 UP000000311 UP000092443 UP000015103 UP000001292 UP000000304 UP000053097 UP000279307
Pfam
PF14736 N_Asn_amidohyd
Interpro
IPR026750
NTAN1
ProteinModelPortal
H9JR94
A0A2A4J421
A0A2H1VA15
A0A212F420
A0A437BJK9
A0A194PEQ9
+ More
A0A194R3S5 D6WDI5 A0A2J7RQ33 A0A1L8E2M8 U5ESL8 A0A1Y1LRQ0 A0A084WQY8 A0A1Q3FH91 A0A182QEE0 A0A182NA40 A0A182PJG6 A0A182R2Y2 A0A182KKD7 A0A182WFU7 A0A182WUM1 A0A182V0C7 B0XAF5 A0A182H4E5 Q17BH8 A0A023EQG7 A0A182JJI4 A0A1Q3FHQ3 A0A2M4CV54 A0A2M4ASY3 A0A2P8Y3E9 A0A182FKE9 A0A2M3Z9R4 A0A2A3E1N8 A0A0L7R0I2 A0A088AP41 A0A0M9A056 E2BEF3 A0A0A1XAY0 K7IPK1 A0A0C9R7S0 A0A232FBE9 T1PKI9 A0A067R6F7 W8C2F4 N6U2W7 J3JUB2 A0A0K8TXW5 A0A034W0Q6 A0A151JVW1 A0A1I8NC15 A0A151WZJ2 E9INW6 F4WH33 A0A195BT04 A0A158NT88 A0A195E355 B4J7N7 A0A154PQR0 B4MRB8 D3TMG3 A0A1A9V8M2 A0A1B6JR27 B4LJ59 B4KPM2 A0A0K8V5E4 A0A034VZ41 A0A1B0G2F3 A0A1I8Q0I7 A0A1I8Q0L4 A0A1A9X2Z3 A0A1B6LLI3 A0A1B6IDW3 A0A0L0CRV6 A0A0P8ZMP3 A0A1W4UM73 A0A1A9ZVI7 A0A0Q9WPI6 A0A3B0J2U3 B4GA64 A0A0R3NLH0 A0A0M4EAG3 A0A0R1DRY6 A0A1B6CM09 A0A0J9RG25 A0A0Q5VKU4 Q7KIS4 E2A596 A0A1W4UMD8 B3MBU5 Q291W5 A0A1A9XAE8 B4P4B3 T1HP56 B4HNZ4 B4QD07 B3NK92 Q8MSD1 A0A026X1Q3 A0A224XVA2
A0A194R3S5 D6WDI5 A0A2J7RQ33 A0A1L8E2M8 U5ESL8 A0A1Y1LRQ0 A0A084WQY8 A0A1Q3FH91 A0A182QEE0 A0A182NA40 A0A182PJG6 A0A182R2Y2 A0A182KKD7 A0A182WFU7 A0A182WUM1 A0A182V0C7 B0XAF5 A0A182H4E5 Q17BH8 A0A023EQG7 A0A182JJI4 A0A1Q3FHQ3 A0A2M4CV54 A0A2M4ASY3 A0A2P8Y3E9 A0A182FKE9 A0A2M3Z9R4 A0A2A3E1N8 A0A0L7R0I2 A0A088AP41 A0A0M9A056 E2BEF3 A0A0A1XAY0 K7IPK1 A0A0C9R7S0 A0A232FBE9 T1PKI9 A0A067R6F7 W8C2F4 N6U2W7 J3JUB2 A0A0K8TXW5 A0A034W0Q6 A0A151JVW1 A0A1I8NC15 A0A151WZJ2 E9INW6 F4WH33 A0A195BT04 A0A158NT88 A0A195E355 B4J7N7 A0A154PQR0 B4MRB8 D3TMG3 A0A1A9V8M2 A0A1B6JR27 B4LJ59 B4KPM2 A0A0K8V5E4 A0A034VZ41 A0A1B0G2F3 A0A1I8Q0I7 A0A1I8Q0L4 A0A1A9X2Z3 A0A1B6LLI3 A0A1B6IDW3 A0A0L0CRV6 A0A0P8ZMP3 A0A1W4UM73 A0A1A9ZVI7 A0A0Q9WPI6 A0A3B0J2U3 B4GA64 A0A0R3NLH0 A0A0M4EAG3 A0A0R1DRY6 A0A1B6CM09 A0A0J9RG25 A0A0Q5VKU4 Q7KIS4 E2A596 A0A1W4UMD8 B3MBU5 Q291W5 A0A1A9XAE8 B4P4B3 T1HP56 B4HNZ4 B4QD07 B3NK92 Q8MSD1 A0A026X1Q3 A0A224XVA2
Ontologies
GO
PANTHER
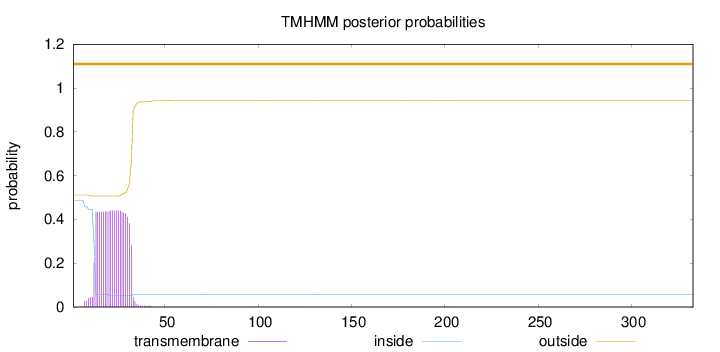
Topology
Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.05822999999999
Exp number, first 60 AAs:
9.0255
Total prob of N-in:
0.48795
outside
1 - 333
Population Genetic Test Statistics
Pi
21.655931
Theta
21.581029
Tajima's D
-1.3532
CLR
1.715863
CSRT
0.079646017699115
Interpretation
Uncertain
