Pre Gene Modal
BGIBMGA012099
Annotation
PREDICTED:_sperm_motility_kinase_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.186
Sequence
CDS
ATGAACAATGGGACGAGACGAGCCTTGACCGGCTCCATTCACAAAATTAGAGAATTTGAACTGGAAAAAGTGAGTTTGGTTGAAGAATTCGATATATTACAAATCGTTGGTGAGGGATGGTTTGGAAAGATTCTTCTTGTTGAACACAAAGCATCAGACACTGAAATTGTGCTGAAAGCGTTGCCTAAACCCTATGTCTCTATACGGGACTTTTACAGAGAGTTCCATTACAGCCTCCACTTAGGTGCGCACCGGAATATTGTGACGACATTTGATGTGGCTTTCGAAACAGCTGGTTTTTATGTGTTTTGCCAAGAATATGCGCCACTAGGTGATCTTACATCTAACATGTTAGATACGGGTATAGGTGAAATCCATTCAAAGAGGGTGGCAAAACAATTGGCTGCAGCATTAGACCATATTCACCAAAGAGAACTAGTTCATAGAGACGTTAAACTAGATAACATATTAATATTCAAATCAGATTTTTCTAGAATTAAGCTATGCGATTTCGGAGAAACGAGGAAGGTTAATGCCGTGGTTAGAAGACGCAATGAATGGTTGCCGTATTCCCCGCCAGAAGTGTTAAAACTATCAATGGACAGCAGCTACAAAGCATTAATTACCCACGATGTTTGGCAATTCGGTATTGTTTTATTTATTTGCCTCACCGGCTGCCTGCCTTGGCAGAAAGCTGCTTTCGACGACCCCCGCTACACGAGCTATTACAATTGGTACAACAGTTCGAATCCATTAAAGAAACAACCGAAGCTATGGAAAATGGTTTCCTCTAGAGCGCAGAGGATGTTTAAAAAGTTCGTTGAACCCAGAGAAGAGAAGAGGGTTAATAATCTACTAGAACTGAATCGATATTTAGATGATCGGTGGCTTGCGAAAGGATACACAGACAGAGTCGCCGGTGAAGACATAGACGAACTCTGTCCATCCATGTACAGTTTTCACAGCGACCCGAATGAGAAAAACATACTGCTCCAACATTTTAGGCACAATGGCCTCGAGACAACTGTAGACAGACAAGCCAAAAAACAAAGAATACGAGACTGGATACAAAATAGTGTCATAGAAGAAGCCGATGAAGAGGAAGATGCATATGAGCCTAGCGTGTACGAAGACAGCGATATAGAACTTGACCACAGACCAAGGAACTACCCGATAGACGATTCCCACAGAGTAACTCTTCGGTACGACACCGAAAAGCATATCAACCCAAGAACCGGCGAAGTCATCGAAGGAGCGAAACCTTCAGCTCCCGAAAAAAAACCTCTATCGTACGACGTTCAACATATTGAGCCCGTGGCCCCGACCGATGTCGTGAGGCGATACGGCCAAAATGACGCATCGCAAAGCTCAACAAAAGACAGCGCTTACGGATCCATGGAAGTTAAAGCAAAAAGAAACGCATACAAAAACAATCAAAAACATTCTACATCTCTCGAATACGGTATAAATAATCCTCCGAGTAGATCGCTAAGTGCCACCTCTATACAACAAAATAAGTCGCCAATCACAAGCCACGCTCAAAGGTTTCAAAGGAGCGAAGAAGTCTATTCAAAGGAATTGGAAAATTTAAAAGGAAGCTCATCGACTCTGCATTCTTTGGCGCACTCGTCTAGATCTAACAGCATCCAAGCGACACACAGCAGCTCCTTCGATAGTCCCACAATCGTAGGCAATAATGTTGCTACACAAAATGAAAGTCCAGTATTATCTTCTACTCATAACAGCAGCTTTGAGAAGCAACAGAATGGCTTAATTCCTCCAAATTCAACGGTGCCACAGCACAGGCCGTCGTATGTATCAATGAAAAGCACTCCTTACGACAATGTTTCGTTCAAAACTGAAAATTCTGATGGAAAACCTGAAACCTCGACGGTTAATGGGAAAAACGAGGAAAACAAGGTTAGTCCCTATTCATCAATGGTGAATATAGTACAAGGAAAAATGAAAATAAATCCCCATCAAGATGATGCCAATAAAAATGCGATAAGCGTTTCAGTGACGGCAGTCAAACAAAACAAAACAGGGAGAAAGAAATCATGA
Protein
MNNGTRRALTGSIHKIREFELEKVSLVEEFDILQIVGEGWFGKILLVEHKASDTEIVLKALPKPYVSIRDFYREFHYSLHLGAHRNIVTTFDVAFETAGFYVFCQEYAPLGDLTSNMLDTGIGEIHSKRVAKQLAAALDHIHQRELVHRDVKLDNILIFKSDFSRIKLCDFGETRKVNAVVRRRNEWLPYSPPEVLKLSMDSSYKALITHDVWQFGIVLFICLTGCLPWQKAAFDDPRYTSYYNWYNSSNPLKKQPKLWKMVSSRAQRMFKKFVEPREEKRVNNLLELNRYLDDRWLAKGYTDRVAGEDIDELCPSMYSFHSDPNEKNILLQHFRHNGLETTVDRQAKKQRIRDWIQNSVIEEADEEEDAYEPSVYEDSDIELDHRPRNYPIDDSHRVTLRYDTEKHINPRTGEVIEGAKPSAPEKKPLSYDVQHIEPVAPTDVVRRYGQNDASQSSTKDSAYGSMEVKAKRNAYKNNQKHSTSLEYGINNPPSRSLSATSIQQNKSPITSHAQRFQRSEEVYSKELENLKGSSSTLHSLAHSSRSNSIQATHSSSFDSPTIVGNNVATQNESPVLSSTHNSSFEKQQNGLIPPNSTVPQHRPSYVSMKSTPYDNVSFKTENSDGKPETSTVNGKNEENKVSPYSSMVNIVQGKMKINPHQDDANKNAISVSVTAVKQNKTGRKKS
Summary
Uniprot
H9JRD9
A0A2A4JA75
A0A2H1VSV8
A0A194R3G0
A0A194PFG4
A0A437BJK6
+ More
A0A0L7LJN7 A0A212FJ77 A0A182PTA7 A0A2C9GUJ1 B4J7H1 A0A084VJU4 B4HT58 Q8SZT8 A0A0B4LFM3 B4QI05 A1ZAH6 A0A0Q5VY65 A0A0R1DZ86 A0A0R3NTW3 A0A0R3NS65 B3NPH3 B4P655 A0A3B0JLD5 Q291U4 A0A0R1DSP3 A0A0Q5W0Y2 A0A1W4UKQ4 B4GA92 A0A3B0J2S7 A0A1W4UYG4 A0A3B0JDX7 A0A0J9RDY4
A0A0L7LJN7 A0A212FJ77 A0A182PTA7 A0A2C9GUJ1 B4J7H1 A0A084VJU4 B4HT58 Q8SZT8 A0A0B4LFM3 B4QI05 A1ZAH6 A0A0Q5VY65 A0A0R1DZ86 A0A0R3NTW3 A0A0R3NS65 B3NPH3 B4P655 A0A3B0JLD5 Q291U4 A0A0R1DSP3 A0A0Q5W0Y2 A0A1W4UKQ4 B4GA92 A0A3B0J2S7 A0A1W4UYG4 A0A3B0JDX7 A0A0J9RDY4
Pubmed
EMBL
BABH01016265
NWSH01002231
PCG68871.1
ODYU01004241
SOQ43907.1
KQ460779
+ More
KPJ12343.1 KQ459606 KPI91763.1 RSAL01000045 RVE50620.1 JTDY01000817 KOB75768.1 AGBW02008303 OWR53782.1 AXCM01000530 CH916367 EDW01095.1 ATLV01013882 KE524908 KFB38238.1 CH480816 EDW48159.1 AE013599 AY070513 AAF57985.2 AAL47984.1 AHN56288.1 CM000362 EDX07376.1 AAO41369.1 AAO41370.1 CH954179 KQS62446.1 CM000158 KRK00199.1 CM000071 KRT01798.1 KRT01797.1 EDV55740.1 EDW91905.1 OUUW01000001 SPP73461.1 EAL25018.1 KRK00200.1 KRK00201.1 KQS62444.1 KQS62445.1 KQS62447.1 KQS62448.1 CH479181 EDW31844.1 SPP73462.1 SPP73460.1 CM002911 KMY94288.1
KPJ12343.1 KQ459606 KPI91763.1 RSAL01000045 RVE50620.1 JTDY01000817 KOB75768.1 AGBW02008303 OWR53782.1 AXCM01000530 CH916367 EDW01095.1 ATLV01013882 KE524908 KFB38238.1 CH480816 EDW48159.1 AE013599 AY070513 AAF57985.2 AAL47984.1 AHN56288.1 CM000362 EDX07376.1 AAO41369.1 AAO41370.1 CH954179 KQS62446.1 CM000158 KRK00199.1 CM000071 KRT01798.1 KRT01797.1 EDV55740.1 EDW91905.1 OUUW01000001 SPP73461.1 EAL25018.1 KRK00200.1 KRK00201.1 KQS62444.1 KQS62445.1 KQS62447.1 KQS62448.1 CH479181 EDW31844.1 SPP73462.1 SPP73460.1 CM002911 KMY94288.1
Proteomes
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JRD9
A0A2A4JA75
A0A2H1VSV8
A0A194R3G0
A0A194PFG4
A0A437BJK6
+ More
A0A0L7LJN7 A0A212FJ77 A0A182PTA7 A0A2C9GUJ1 B4J7H1 A0A084VJU4 B4HT58 Q8SZT8 A0A0B4LFM3 B4QI05 A1ZAH6 A0A0Q5VY65 A0A0R1DZ86 A0A0R3NTW3 A0A0R3NS65 B3NPH3 B4P655 A0A3B0JLD5 Q291U4 A0A0R1DSP3 A0A0Q5W0Y2 A0A1W4UKQ4 B4GA92 A0A3B0J2S7 A0A1W4UYG4 A0A3B0JDX7 A0A0J9RDY4
A0A0L7LJN7 A0A212FJ77 A0A182PTA7 A0A2C9GUJ1 B4J7H1 A0A084VJU4 B4HT58 Q8SZT8 A0A0B4LFM3 B4QI05 A1ZAH6 A0A0Q5VY65 A0A0R1DZ86 A0A0R3NTW3 A0A0R3NS65 B3NPH3 B4P655 A0A3B0JLD5 Q291U4 A0A0R1DSP3 A0A0Q5W0Y2 A0A1W4UKQ4 B4GA92 A0A3B0J2S7 A0A1W4UYG4 A0A3B0JDX7 A0A0J9RDY4
PDB
2H9V
E-value=3.94822e-17,
Score=218
Ontologies
KEGG
GO
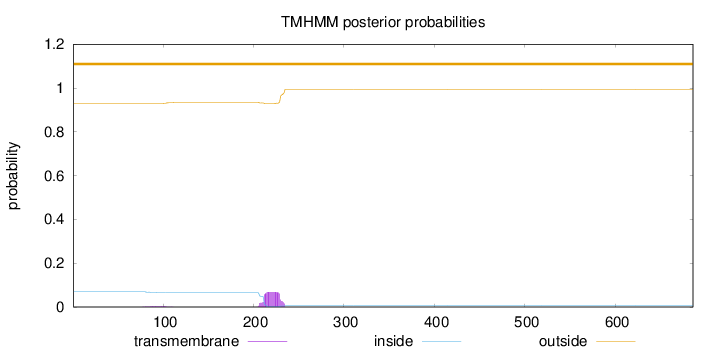
Topology
Length:
686
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.49989
Exp number, first 60 AAs:
0.00056
Total prob of N-in:
0.06990
outside
1 - 686
Population Genetic Test Statistics
Pi
5.304069
Theta
14.20672
Tajima's D
-1.210409
CLR
13.967917
CSRT
0.0987450627468627
Interpretation
Uncertain
