Pre Gene Modal
BGIBMGA012053
Annotation
PREDICTED:_putative_sodium-coupled_neutral_amino_acid_transporter_10_isoform_X1_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 2.28
Sequence
CDS
ATGGGAGTCGCTGGACAATCTATAACGCTGGCAAATAGCATTATAGGCGTTGGCATATTAGCCATGCCGTATTGCTTTCAACAATGTGGTATATTGCTGGCAACATTAATCTTGCTAACCATGGGCTTAGTTTCGAGACTTTGCTGCTATTTCCTTTTGAAATCTGCTTTGTTGGCCCGAAGGAGAAACTTTGAATTTTTAGCGTTTCACGTATTCGGACCGGCTGGGAAGATGGCCGTTGAAATCGGTATTATCGGATTTCTGATGGGAACGTGCATAGCTTACTTCGTGGTAGTCGGAGATTTAGGGCCGCAGATAATTTCAAAAATGTTGAACGTTAACCAAAGCGATATATTGAGGACATCGATAATGGTTGTGGTGTCGCTAGTGTGCGTGCTGCCACTCGGCTTGCTCCGTAACGTCGACAGTCTCAGTAACGTGAGCGCAGCCACCATATGCTTCTATCTCTGTCTCGTCGTGAAGGTGATAACGGAAGCGACGTCACAGTTGTTCGAAGAGGAATTACAGAATCGTATGGAGCTGTGGAAGCCGTCCGGGGTGCTTCAGTGCGTCCCGATATTCTCGATGGCGCTGTTTTGTCAAACCCAGCTATTCGAAATATTCGAGTCGCTGCCGACCCTGTCCTTAGAGAAGATGAATTTGGTTACAAAGAATGCGATAAACATTTGTACAGGAGTCTATTTCACTCTAGGTCTGTTCGGATACATTGCGTTCTGCTCACAGGATATATCCGGTAACATCTTGATGAGCCTAAGTCCGACGATGGCGAGTGACGTCATCAAGCTCGGGTTCGTGATGTCGCTCGCGTTCAGTTTCCCGCTGATCATATTCCCGTGTCGGGCGAGCCTCTATTCGTTCCTGTACAAAAAGGTCCACTCGTCCCACCACGACCACATGATCAACCATTCGATCCCGCAGACCACGTTCCGATGCATAACGGTGGGCATTATAGGCGTGGCTCTGGTCGTGGGTCTGCTGGTCCCCAACATCGAGCTGGTGTTGGGGCTGGCGGGCTCCACCATCGGCGTGCTGGTCTGCGTGGTATTCCCGGCCGCTTGCTTCGTCAACGTCACCTTCAAGAATACGAACGAGAGGATCCTGGCCAAGGGCATTATCGTCCTTGGATTAATTATAATGGTACTCGGGACATACGCTAACTTGCAAGCCGCTGAAGGCAAACACGAGAGATACGACGAGAAATACATCACACAAGAAAAGATTGATAAAATGGTTGAGGACTTCTTCCAGAAGAGAGAGAAATTGGAGAAAATAATCCCAGCCGCCGAGGGTGAGATTCTTCCAGATGACGCACAAAAAATAATCGAAAGTAACATAATAAGGGAATCGGAGGTTCAACCCCCTAATCCTGTACCTCCAGACTCATCCAAAGAAAAAATCCCAGATATAGCTTTAATTGAAAACAAAAAAGTCCAAGAAGCTGTGTCCGATGTGATCAAAGTGAACGGCGATAAAGAACCGGAACTACATCCCAAAGACGTAAAATCCAAACTAAACGAAGTCAAAGCCGTGATCGACGGTAATGTTATGAAAGAAGAAATAGTTAAGAAAATAGAAGAACCTAAACAGATGAACGAAAACGAAGATAGATTGCGCATACTGAAACAACAGAAACTTATCGAAACAATCAAGAAGCACGGCGAAGAACAAAAAGAACTAGTTCAGGAGCAAAAGGAAATTATAGATGAGTTACTGAAGAACAAGAAGGAAGAGAAAGAAAACAGCAATGATGGTATAAAGTTGTCGGAAGTGAAAGGGATAGCGGATGTACCTCCAGGACGTCCGGCTGGTATCCCCGAGACGCACGCCGCTCTACAGAACGCTGAGGAGTCGCCAAACGAAGCTAAACAACAAATAGACCAAAGAGCAGCTGAAGCCATTAAAGTTATCGAGGAAGTTATTAAACCAGAGCAACCAAAGAACATACGAAATGTAGCCGACAATGTTAACTCTGAACATATACAACAAAATGCACAAAACGCTAATCCTTCTAACAATCCCAACGCAGATATCGGAAAGTTAAAAGAAATCTCCCGGATTCCGGAACCGATTGTCAATGAATTGAACAACTTCAACAGACCGTTGCCCGTTAACGTTCGATCCGCAGTTAAACAAGAATCACCACCGCAAGTCGGTCAAGGCGACGTCAACAAAGAAACACAGAAACAAAACTTAGATGATACCGAAATAAAAAAACTCCAAGAAGCTAGTGTTCAAAAAAATATACGAACACTAAACTCGGATTCACCAGATGTGCCTCAGTCGTACAAACAAGGAATGGATAATAAAAACGTAATGCAAAACGTACCATTGGCTCTAGCGATGAACGACAAACAGTTAAAGAATGAAATCAACAATGGCGTACCAAAGAAAAATGTAGAGAAACTCAGTAATGATATCGTTGACCAACAAAACCCGAATCTCGTTCCGATGCGACAGAAGAGGCAAACTATAGACTGTTCGAATAAAGCCACTTTGCGAGTGGAGGACAGAAGGATATGTGAAAATTTAATTGGATATCCAAATAGCAACGACGTAATTTTACCCGAAGTCGATTTGAACGACGCCTTCGTTAAACGTATACCTATCGACGAAAACGTTTTACGCGTCGGTAGGTCCTTGAAAATATACGACGAGAGAACGAAGGAGATGAAAGAGAGATAG
Protein
MGVAGQSITLANSIIGVGILAMPYCFQQCGILLATLILLTMGLVSRLCCYFLLKSALLARRRNFEFLAFHVFGPAGKMAVEIGIIGFLMGTCIAYFVVVGDLGPQIISKMLNVNQSDILRTSIMVVVSLVCVLPLGLLRNVDSLSNVSAATICFYLCLVVKVITEATSQLFEEELQNRMELWKPSGVLQCVPIFSMALFCQTQLFEIFESLPTLSLEKMNLVTKNAINICTGVYFTLGLFGYIAFCSQDISGNILMSLSPTMASDVIKLGFVMSLAFSFPLIIFPCRASLYSFLYKKVHSSHHDHMINHSIPQTTFRCITVGIIGVALVVGLLVPNIELVLGLAGSTIGVLVCVVFPAACFVNVTFKNTNERILAKGIIVLGLIIMVLGTYANLQAAEGKHERYDEKYITQEKIDKMVEDFFQKREKLEKIIPAAEGEILPDDAQKIIESNIIRESEVQPPNPVPPDSSKEKIPDIALIENKKVQEAVSDVIKVNGDKEPELHPKDVKSKLNEVKAVIDGNVMKEEIVKKIEEPKQMNENEDRLRILKQQKLIETIKKHGEEQKELVQEQKEIIDELLKNKKEEKENSNDGIKLSEVKGIADVPPGRPAGIPETHAALQNAEESPNEAKQQIDQRAAEAIKVIEEVIKPEQPKNIRNVADNVNSEHIQQNAQNANPSNNPNADIGKLKEISRIPEPIVNELNNFNRPLPVNVRSAVKQESPPQVGQGDVNKETQKQNLDDTEIKKLQEASVQKNIRTLNSDSPDVPQSYKQGMDNKNVMQNVPLALAMNDKQLKNEINNGVPKKNVEKLSNDIVDQQNPNLVPMRQKRQTIDCSNKATLRVEDRRICENLIGYPNSNDVILPEVDLNDAFVKRIPIDENVLRVGRSLKIYDERTKEMKER
Summary
Uniprot
EMBL
Proteomes
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
Ontologies
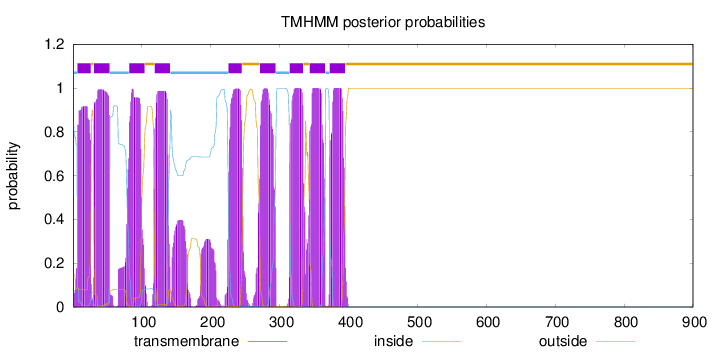
Topology
Length:
900
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
208.73878
Exp number, first 60 AAs:
41.20816
Total prob of N-in:
0.85306
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 30
TMhelix
31 - 53
inside
54 - 81
TMhelix
82 - 104
outside
105 - 118
TMhelix
119 - 141
inside
142 - 225
TMhelix
226 - 245
outside
246 - 271
TMhelix
272 - 294
inside
295 - 314
TMhelix
315 - 334
outside
335 - 343
TMhelix
344 - 366
inside
367 - 372
TMhelix
373 - 395
outside
396 - 900
Population Genetic Test Statistics
Pi
0.209853
Theta
1.906146
Tajima's D
-2.042684
CLR
555.642063
CSRT
0.00509974501274936
Interpretation
Possibly Positive selection
