Gene
KWMTBOMO06817
Pre Gene Modal
BGIBMGA012050
Annotation
PREDICTED:_uncharacterized_protein_LOC101741648_isoform_X1_[Bombyx_mori]
Full name
Mitochondrial cardiolipin hydrolase
Alternative Name
Choline phosphatase 6
Mitochondrial phospholipase
Phosphatidylcholine-hydrolyzing phospholipase D6
Phospholipase D6
Protein zucchini homolog
Mitochondrial phospholipase
Phosphatidylcholine-hydrolyzing phospholipase D6
Phospholipase D6
Protein zucchini homolog
Location in the cell
Cytoplasmic Reliability : 2.206
Sequence
CDS
ATGCCTGGGATACACAATGTAGCAATACAAGGACGCTTAGTTGAAATGCTGAAAAAGAAAAATATCAAGATTTCAATTGTAATAGATCAGTCCGGGTGCAACGAACCAAATGATGTCTTCATAAAGGAACTTTTAGATGCTGGTGCAGTAATCAAGTACATTAATACAGAACCGTATACAATGCAACATAAATTCTGTTTGATCGACGACAAAGTTTTAATGACAGGTACACTAAACTGGGGAAACGATCGATCGTCAGATCACTGGAACTACGTCTACATTACAAGCAAGCCAAAGCTAGTTGAACCAGTCAGCAAAGAGTTCAAATATATGTGGCTCAGTTCAAAAGATTTGAACTATTCCCAGTTTGATAGCAAACCTATCAACGAAGATGAGTGTTGTAACACAGAAGTCGTTAATCTTACAGAAAACTATAAATCAGTCCCGTTTGGAAATAAAGAGACGCTGACTTCTGAAATATGCATAGTGTAA
Protein
MPGIHNVAIQGRLVEMLKKKNIKISIVIDQSGCNEPNDVFIKELLDAGAVIKYINTEPYTMQHKFCLIDDKVLMTGTLNWGNDRSSDHWNYVYITSKPKLVEPVSKEFKYMWLSSKDLNYSQFDSKPINEDECCNTEVVNLTENYKSVPFGNKETLTSEICIV
Summary
Description
Endonuclease that plays a critical role in PIWI-interacting RNA (piRNA) biogenesis during spermatogenesis. piRNAs provide essential protection against the activity of mobile genetic elements (By similarity). piRNA-mediated transposon silencing is thus critical for maintaining genome stability, in particular in germline cells when transposons are mobilized as a consequence of wide-spread genomic demethylation (By similarity). Has been proposed to act as a cardiolipin hydrolase to generate phosphatidic acid at mitochondrial surface (By similarity). Although it cannot be excluded that it can act as a phospholipase in some circumstances, it should be noted that cardiolipin hydrolase activity is either undetectable in vitro, or very low (PubMed:21397848). In addition, cardiolipin is almost exclusively found on the inner mitochondrial membrane, while PLD6 localizes to the outer mitochondrial membrane, facing the cytosol (PubMed:21397848). Has been shown to be a backbone-non-specific, single strand-specific nuclease, cleaving either RNA or DNA substrates with similar affinity. Produces 5' phosphate and 3' hydroxyl termini, suggesting it could directly participate in the processing of primary piRNA transcripts (By similarity). Also acts as a regulator of mitochondrial shape through facilitating mitochondrial fusion (PubMed:17028579, PubMed:26711011).
Subunit
Homodimer (PubMed:17028579). Interacts with MOV10L1 (By similarity). Interacts with MIGA1 and MIGA2; possibly facilitating homodimer formation (PubMed:26711011).
Similarity
Belongs to the phospholipase D family. MitoPLD/Zucchini subfamily.
Keywords
Complete proteome
Differentiation
Endonuclease
Hydrolase
Lipid degradation
Lipid metabolism
Meiosis
Membrane
Metal-binding
Mitochondrion
Mitochondrion outer membrane
Nuclease
Polymorphism
Reference proteome
Spermatogenesis
Transmembrane
Transmembrane helix
Zinc
Zinc-finger
Feature
chain Mitochondrial cardiolipin hydrolase
sequence variant In dbSNP:rs17856924.
sequence variant In dbSNP:rs17856924.
Uniprot
EC Number
3.1.-.-
Pubmed
EMBL
BABH01016273
KQ460779
KPJ12352.1
KQ459606
KPI91770.1
AGBW02014707
+ More
OWR41151.1 ODYU01005213 SOQ45851.1 GAIX01012278 JAA80282.1 JTDY01007674 KOB65035.1 RSAL01000045 RVE50614.1 GFAC01001823 JAT97365.1 GBBM01003377 JAC32041.1 AK090899 AC055811 BC031263 CABD030039199 CP026250 AWP05796.1 AWP05797.1 AWP05798.1 AWP05799.1 AJFE02026409 AACZ04059834 AK305029 NBAG03000529 BAK62023.1 PNI17172.1 CP013118 ALO17377.1 KN122870 KFO27648.1 AERX01018863 JH173139 EHB15943.1 QUSF01011075 RLV62105.1 ADFV01121093
OWR41151.1 ODYU01005213 SOQ45851.1 GAIX01012278 JAA80282.1 JTDY01007674 KOB65035.1 RSAL01000045 RVE50614.1 GFAC01001823 JAT97365.1 GBBM01003377 JAC32041.1 AK090899 AC055811 BC031263 CABD030039199 CP026250 AWP05796.1 AWP05797.1 AWP05798.1 AWP05799.1 AJFE02026409 AACZ04059834 AK305029 NBAG03000529 BAK62023.1 PNI17172.1 CP013118 ALO17377.1 KN122870 KFO27648.1 AERX01018863 JH173139 EHB15943.1 QUSF01011075 RLV62105.1 ADFV01121093
Proteomes
Pfam
PF13091 PLDc_2
ProteinModelPortal
PDB
4GGK
E-value=6.94311e-08,
Score=131
Ontologies
GO
Topology
Subcellular location
Mitochondrion outer membrane
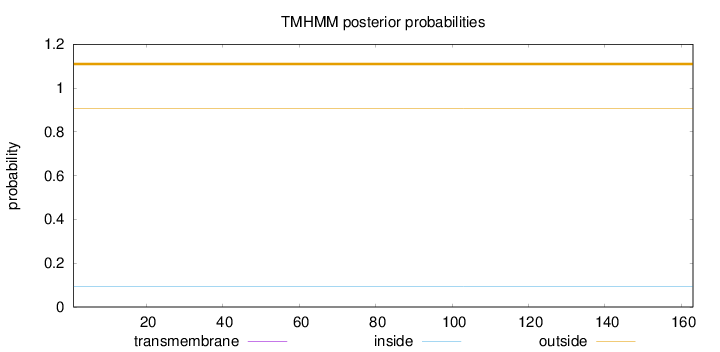
Length:
163
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09364
outside
1 - 163
Population Genetic Test Statistics
Pi
12.397678
Theta
15.929056
Tajima's D
-0.997964
CLR
7.904372
CSRT
0.137093145342733
Interpretation
Uncertain
