Gene
KWMTBOMO06807 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012106
Annotation
PREDICTED:_tyrosine-protein_phosphatase_Lar_isoform_X1_[Plutella_xylostella]
Full name
Tyrosine-protein phosphatase Lar
Alternative Name
Protein-tyrosine-phosphate phosphohydrolase
dLAR
dLAR
Location in the cell
Extracellular Reliability : 3.268
Sequence
CDS
ATGGATAAACCAGCTGGATTAATTTCCTCATTGGTGGTTTGCATTCTGCTACTGCAAGGGCTAGTCGAAAGTGCGCCATCATCTGACGCTCATCTCTTTCCACCTCCTCCAATTGCATATGTCGCTCGGGGAGGCACCGTAACATTAAAATGCTCCGCAACCGGCTCCCCGACCCCTACGATTAGATGGAAGAAAGGATCAGAATGGCTGACTCCCGAAGACAATACAGTGTCAAAGCATGTTCTGACACTAACAAACGTTACAGAAGAAGGTAAATACACATGTGTCTTTAAAACGGGAGAGCTTGTTTCTTCAGTGACGACCGCTGTTAAAATCAGACCATTACCACGCACACCGCACAGCGTCCATATGTCAGACGTCACGGACACCAGCGCTCAGTTGGAATGGTCGTATTCCTACATCCCAGGGACAGACGATCCAACTCATTACCACTTGCTTTTGAAACCTAAGCAACAAAACATTGAACCTCAGGATATAACAACAGGGACGAGAACAAGTTTCGAACTGACAAACTTGACTCCGGCAACCGAATATCAAGTTCAGGTCATTGCAGCTGGTACTGTCGGTAAAGGACCCGCCACAGAACCAGTTTTCTTTACTACCAAATTTTAA
Protein
MDKPAGLISSLVVCILLLQGLVESAPSSDAHLFPPPPIAYVARGGTVTLKCSATGSPTPTIRWKKGSEWLTPEDNTVSKHVLTLTNVTEEGKYTCVFKTGELVSSVTTAVKIRPLPRTPHSVHMSDVTDTSAQLEWSYSYIPGTDDPTHYHLLLKPKQQNIEPQDITTGTRTSFELTNLTPATEYQVQVIAAGTVGKGPATEPVFFTTKF
Summary
Description
Possible cell adhesion receptor (Probable). It possesses an intrinsic protein tyrosine phosphatase activity (PTPase) (PubMed:2554325). It controls motor axon guidance (PubMed:8598047). In the developing eye, has a role in normal axonal targeting of the R7 photoreceptor, where it negatively regulates bdl (PubMed:24174674). Inhibits bdl cell adhesion activity in vitro; this effect is independent of its PTPase function (PubMed:24174674).
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Similarity
Belongs to the protein-tyrosine phosphatase family. Receptor class 2A subfamily.
Keywords
3D-structure
Cell adhesion
Complete proteome
Disulfide bond
Glycoprotein
Heparin-binding
Hydrolase
Immunoglobulin domain
Membrane
Neurogenesis
Phosphoprotein
Protein phosphatase
Receptor
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Feature
chain Tyrosine-protein phosphatase Lar
Uniprot
H9JRE6
A0A212ET51
A0A1B0A8A2
A0A195BU66
A0A195F635
A0A151IC73
+ More
A0A158NJY1 A0A2H1W025 A0A195EJ36 B4IN44 A0A1B0BHE1 A0A194R538 A0A310SFZ7 A0A1B0G0C1 A0A3S2P2E0 A0A1A9V1C7 A0A1B6LMQ6 A0A026VZB6 A0A0M9AAH8 A0A3L8DRD2 A0A194PER9 E0V9F8 A0A182GI36 E2B1N6 B0X1M9 A0A0L0C2Q5 A0A1A9WX66 A0A1I8MPV9 A0A0P8XW02 A0A0P8Y3Z8 A0A0P8XG08 A0A0P9A4T0 A0A0N8NZ76 A0A0P9BNX7 B3ML69 A0A0P8XF82 A0A0P9BNY8 A0A2A3EPX0 A0A0A1WP55 A0A0P8XFK7 E2BS15 A0A034W7Z9 A0A034W6H2 A0A1I8NX70 A0A1I8NX96 A0A1I8NX65 A0A1D2ND36 D6WFC0 A0A182P152 A0A182KAD5 A0A139WLZ0 F4WXQ1 A0A1A9X874 A0A1Y1MEI9 A0A182R1N6 A0A2M4ATV6 Q17AI9 A0A0T6B9R3 M9PG93 M9PDF7 A0A0R1DTR4 A0A0R1DVT1 B4PB20 A0A0R1DVC6 A0A0R1DQC6 A0A0R1DVI0 A0A0Q9X675 A0A0R1DPG7 M9NCY3 A0A0Q9X679 A0A182XH68 A0A0Q9XFR8 A0A1W4V7C4 A0A0Q9XJC1 A0A0Q9XG67 A0A1W4UUY1 B4KIB2 A0A1W4V887 A0A0J9R4Q9 A0A1W4V7B8 A0A1W4V6V6 A0A1W4V6W3 M9PDR3 A0A0Q5VXV8 A0A1W4UUX7 A0A0Q5VJF1 A0A0J9R470 A8DZ18 A0A0J9R504 A0A0Q9XHM4 A0A0Q5VW78 A0A0J9TRP5 P16621 A0A0J9R500 A0A087ZS62 A0A0Q5VZ03 A0A0Q5VJR8 M9PD76 A0A0Q5VKK9 B4QA19
A0A158NJY1 A0A2H1W025 A0A195EJ36 B4IN44 A0A1B0BHE1 A0A194R538 A0A310SFZ7 A0A1B0G0C1 A0A3S2P2E0 A0A1A9V1C7 A0A1B6LMQ6 A0A026VZB6 A0A0M9AAH8 A0A3L8DRD2 A0A194PER9 E0V9F8 A0A182GI36 E2B1N6 B0X1M9 A0A0L0C2Q5 A0A1A9WX66 A0A1I8MPV9 A0A0P8XW02 A0A0P8Y3Z8 A0A0P8XG08 A0A0P9A4T0 A0A0N8NZ76 A0A0P9BNX7 B3ML69 A0A0P8XF82 A0A0P9BNY8 A0A2A3EPX0 A0A0A1WP55 A0A0P8XFK7 E2BS15 A0A034W7Z9 A0A034W6H2 A0A1I8NX70 A0A1I8NX96 A0A1I8NX65 A0A1D2ND36 D6WFC0 A0A182P152 A0A182KAD5 A0A139WLZ0 F4WXQ1 A0A1A9X874 A0A1Y1MEI9 A0A182R1N6 A0A2M4ATV6 Q17AI9 A0A0T6B9R3 M9PG93 M9PDF7 A0A0R1DTR4 A0A0R1DVT1 B4PB20 A0A0R1DVC6 A0A0R1DQC6 A0A0R1DVI0 A0A0Q9X675 A0A0R1DPG7 M9NCY3 A0A0Q9X679 A0A182XH68 A0A0Q9XFR8 A0A1W4V7C4 A0A0Q9XJC1 A0A0Q9XG67 A0A1W4UUY1 B4KIB2 A0A1W4V887 A0A0J9R4Q9 A0A1W4V7B8 A0A1W4V6V6 A0A1W4V6W3 M9PDR3 A0A0Q5VXV8 A0A1W4UUX7 A0A0Q5VJF1 A0A0J9R470 A8DZ18 A0A0J9R504 A0A0Q9XHM4 A0A0Q5VW78 A0A0J9TRP5 P16621 A0A0J9R500 A0A087ZS62 A0A0Q5VZ03 A0A0Q5VJR8 M9PD76 A0A0Q5VKK9 B4QA19
EC Number
3.1.3.48
Pubmed
19121390
22118469
21347285
17994087
26354079
24508170
+ More
30249741 20566863 26483478 20798317 26108605 25315136 18057021 25830018 25348373 27289101 18362917 19820115 21719571 28004739 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 2554325 8598047 12537569 18327897 24174674 21402080
30249741 20566863 26483478 20798317 26108605 25315136 18057021 25830018 25348373 27289101 18362917 19820115 21719571 28004739 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 2554325 8598047 12537569 18327897 24174674 21402080
EMBL
BABH01016295
AGBW02012669
OWR44621.1
KQ976403
KYM92154.1
KQ981805
+ More
KYN35529.1 KQ978064 KYM97687.1 ADTU01002015 ADTU01002016 ADTU01002017 ADTU01002018 ADTU01002019 ADTU01002020 ADTU01002021 ADTU01002022 ADTU01002023 ADTU01002024 ODYU01005213 SOQ45854.1 KQ978801 KYN28275.1 CH481513 EDW55890.1 JXJN01014297 JXJN01014298 KQ460779 KPJ12355.1 KQ760382 OAD60720.1 CCAG010016271 RSAL01000045 RVE50612.1 GEBQ01015049 JAT24928.1 KK107570 EZA48821.1 KQ435714 KOX79189.1 QOIP01000005 RLU22970.1 KQ459606 KPI91772.1 DS234992 EEB10014.1 JXUM01012059 JXUM01012060 JXUM01012061 KQ560344 KXJ82941.1 GL444953 EFN60392.1 DS232264 EDS38757.1 JRES01000979 KNC26541.1 CH902620 KPU73545.1 KPU73551.1 KPU73550.1 KPU73547.1 KPU73548.1 KPU73543.1 EDV31687.1 KPU73544.1 KPU73549.1 KPU73552.1 KZ288204 PBC33179.1 GBXI01013650 JAD00642.1 KPU73546.1 GL450107 EFN81531.1 GAKP01009044 JAC49908.1 GAKP01009045 JAC49907.1 LJIJ01000090 ODN02995.1 KQ971319 EFA00482.2 KYB28847.1 GL888427 EGI60979.1 GEZM01033883 JAV84073.1 GGFK01010697 MBW44018.1 CH477334 EAT43263.1 LJIG01002886 KRT84054.1 AE014134 AGB93132.1 AGB93131.1 CM000158 KRJ99186.1 KRJ99187.1 EDW90449.1 KRJ99184.1 KRJ99182.1 KRJ99185.1 CH933807 KRG03743.1 KRJ99181.1 KRJ99183.1 AFH03768.1 KRG03744.1 KRG03746.1 KRG03747.1 KRG03742.1 EDW13409.2 CM002910 KMY91068.1 AGB93130.1 CH954179 KQS61874.1 KQS61869.1 KMY91069.1 KMY91072.1 AAN11057.2 KMY91071.1 KRG03745.1 KQS61870.1 KMY91070.1 M27700 U36857 U36849 U36850 U36851 U36852 U36853 U36854 U36855 U36856 AY051985 KMY91066.1 KQS61871.1 KQS61872.1 AGB93129.1 KQS61873.1 CM000361 EDX05549.1 KMY91064.1
KYN35529.1 KQ978064 KYM97687.1 ADTU01002015 ADTU01002016 ADTU01002017 ADTU01002018 ADTU01002019 ADTU01002020 ADTU01002021 ADTU01002022 ADTU01002023 ADTU01002024 ODYU01005213 SOQ45854.1 KQ978801 KYN28275.1 CH481513 EDW55890.1 JXJN01014297 JXJN01014298 KQ460779 KPJ12355.1 KQ760382 OAD60720.1 CCAG010016271 RSAL01000045 RVE50612.1 GEBQ01015049 JAT24928.1 KK107570 EZA48821.1 KQ435714 KOX79189.1 QOIP01000005 RLU22970.1 KQ459606 KPI91772.1 DS234992 EEB10014.1 JXUM01012059 JXUM01012060 JXUM01012061 KQ560344 KXJ82941.1 GL444953 EFN60392.1 DS232264 EDS38757.1 JRES01000979 KNC26541.1 CH902620 KPU73545.1 KPU73551.1 KPU73550.1 KPU73547.1 KPU73548.1 KPU73543.1 EDV31687.1 KPU73544.1 KPU73549.1 KPU73552.1 KZ288204 PBC33179.1 GBXI01013650 JAD00642.1 KPU73546.1 GL450107 EFN81531.1 GAKP01009044 JAC49908.1 GAKP01009045 JAC49907.1 LJIJ01000090 ODN02995.1 KQ971319 EFA00482.2 KYB28847.1 GL888427 EGI60979.1 GEZM01033883 JAV84073.1 GGFK01010697 MBW44018.1 CH477334 EAT43263.1 LJIG01002886 KRT84054.1 AE014134 AGB93132.1 AGB93131.1 CM000158 KRJ99186.1 KRJ99187.1 EDW90449.1 KRJ99184.1 KRJ99182.1 KRJ99185.1 CH933807 KRG03743.1 KRJ99181.1 KRJ99183.1 AFH03768.1 KRG03744.1 KRG03746.1 KRG03747.1 KRG03742.1 EDW13409.2 CM002910 KMY91068.1 AGB93130.1 CH954179 KQS61874.1 KQS61869.1 KMY91069.1 KMY91072.1 AAN11057.2 KMY91071.1 KRG03745.1 KQS61870.1 KMY91070.1 M27700 U36857 U36849 U36850 U36851 U36852 U36853 U36854 U36855 U36856 AY051985 KMY91066.1 KQS61871.1 KQS61872.1 AGB93129.1 KQS61873.1 CM000361 EDX05549.1 KMY91064.1
Proteomes
UP000005204
UP000007151
UP000092445
UP000078540
UP000078541
UP000078542
+ More
UP000005205 UP000078492 UP000001292 UP000092460 UP000053240 UP000092444 UP000283053 UP000078200 UP000053097 UP000053105 UP000279307 UP000053268 UP000009046 UP000069940 UP000249989 UP000000311 UP000002320 UP000037069 UP000091820 UP000095301 UP000007801 UP000242457 UP000008237 UP000095300 UP000094527 UP000007266 UP000075885 UP000075881 UP000007755 UP000092443 UP000075900 UP000008820 UP000000803 UP000002282 UP000009192 UP000076407 UP000192221 UP000008711 UP000005203 UP000000304
UP000005205 UP000078492 UP000001292 UP000092460 UP000053240 UP000092444 UP000283053 UP000078200 UP000053097 UP000053105 UP000279307 UP000053268 UP000009046 UP000069940 UP000249989 UP000000311 UP000002320 UP000037069 UP000091820 UP000095301 UP000007801 UP000242457 UP000008237 UP000095300 UP000094527 UP000007266 UP000075885 UP000075881 UP000007755 UP000092443 UP000075900 UP000008820 UP000000803 UP000002282 UP000009192 UP000076407 UP000192221 UP000008711 UP000005203 UP000000304
Interpro
IPR003598
Ig_sub2
+ More
IPR036179 Ig-like_dom_sf
IPR003961 FN3_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR036116 FN3_sf
IPR013098 Ig_I-set
IPR029021 Prot-tyrosine_phosphatase-like
IPR003595 Tyr_Pase_cat
IPR016130 Tyr_Pase_AS
IPR000387 TYR_PHOSPHATASE_dom
IPR000242 PTPase_domain
IPR036179 Ig-like_dom_sf
IPR003961 FN3_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR036116 FN3_sf
IPR013098 Ig_I-set
IPR029021 Prot-tyrosine_phosphatase-like
IPR003595 Tyr_Pase_cat
IPR016130 Tyr_Pase_AS
IPR000387 TYR_PHOSPHATASE_dom
IPR000242 PTPase_domain
Gene 3D
CDD
ProteinModelPortal
H9JRE6
A0A212ET51
A0A1B0A8A2
A0A195BU66
A0A195F635
A0A151IC73
+ More
A0A158NJY1 A0A2H1W025 A0A195EJ36 B4IN44 A0A1B0BHE1 A0A194R538 A0A310SFZ7 A0A1B0G0C1 A0A3S2P2E0 A0A1A9V1C7 A0A1B6LMQ6 A0A026VZB6 A0A0M9AAH8 A0A3L8DRD2 A0A194PER9 E0V9F8 A0A182GI36 E2B1N6 B0X1M9 A0A0L0C2Q5 A0A1A9WX66 A0A1I8MPV9 A0A0P8XW02 A0A0P8Y3Z8 A0A0P8XG08 A0A0P9A4T0 A0A0N8NZ76 A0A0P9BNX7 B3ML69 A0A0P8XF82 A0A0P9BNY8 A0A2A3EPX0 A0A0A1WP55 A0A0P8XFK7 E2BS15 A0A034W7Z9 A0A034W6H2 A0A1I8NX70 A0A1I8NX96 A0A1I8NX65 A0A1D2ND36 D6WFC0 A0A182P152 A0A182KAD5 A0A139WLZ0 F4WXQ1 A0A1A9X874 A0A1Y1MEI9 A0A182R1N6 A0A2M4ATV6 Q17AI9 A0A0T6B9R3 M9PG93 M9PDF7 A0A0R1DTR4 A0A0R1DVT1 B4PB20 A0A0R1DVC6 A0A0R1DQC6 A0A0R1DVI0 A0A0Q9X675 A0A0R1DPG7 M9NCY3 A0A0Q9X679 A0A182XH68 A0A0Q9XFR8 A0A1W4V7C4 A0A0Q9XJC1 A0A0Q9XG67 A0A1W4UUY1 B4KIB2 A0A1W4V887 A0A0J9R4Q9 A0A1W4V7B8 A0A1W4V6V6 A0A1W4V6W3 M9PDR3 A0A0Q5VXV8 A0A1W4UUX7 A0A0Q5VJF1 A0A0J9R470 A8DZ18 A0A0J9R504 A0A0Q9XHM4 A0A0Q5VW78 A0A0J9TRP5 P16621 A0A0J9R500 A0A087ZS62 A0A0Q5VZ03 A0A0Q5VJR8 M9PD76 A0A0Q5VKK9 B4QA19
A0A158NJY1 A0A2H1W025 A0A195EJ36 B4IN44 A0A1B0BHE1 A0A194R538 A0A310SFZ7 A0A1B0G0C1 A0A3S2P2E0 A0A1A9V1C7 A0A1B6LMQ6 A0A026VZB6 A0A0M9AAH8 A0A3L8DRD2 A0A194PER9 E0V9F8 A0A182GI36 E2B1N6 B0X1M9 A0A0L0C2Q5 A0A1A9WX66 A0A1I8MPV9 A0A0P8XW02 A0A0P8Y3Z8 A0A0P8XG08 A0A0P9A4T0 A0A0N8NZ76 A0A0P9BNX7 B3ML69 A0A0P8XF82 A0A0P9BNY8 A0A2A3EPX0 A0A0A1WP55 A0A0P8XFK7 E2BS15 A0A034W7Z9 A0A034W6H2 A0A1I8NX70 A0A1I8NX96 A0A1I8NX65 A0A1D2ND36 D6WFC0 A0A182P152 A0A182KAD5 A0A139WLZ0 F4WXQ1 A0A1A9X874 A0A1Y1MEI9 A0A182R1N6 A0A2M4ATV6 Q17AI9 A0A0T6B9R3 M9PG93 M9PDF7 A0A0R1DTR4 A0A0R1DVT1 B4PB20 A0A0R1DVC6 A0A0R1DQC6 A0A0R1DVI0 A0A0Q9X675 A0A0R1DPG7 M9NCY3 A0A0Q9X679 A0A182XH68 A0A0Q9XFR8 A0A1W4V7C4 A0A0Q9XJC1 A0A0Q9XG67 A0A1W4UUY1 B4KIB2 A0A1W4V887 A0A0J9R4Q9 A0A1W4V7B8 A0A1W4V6V6 A0A1W4V6W3 M9PDR3 A0A0Q5VXV8 A0A1W4UUX7 A0A0Q5VJF1 A0A0J9R470 A8DZ18 A0A0J9R504 A0A0Q9XHM4 A0A0Q5VW78 A0A0J9TRP5 P16621 A0A0J9R500 A0A087ZS62 A0A0Q5VZ03 A0A0Q5VJR8 M9PD76 A0A0Q5VKK9 B4QA19
PDB
4YH7
E-value=6.47821e-21,
Score=245
Ontologies
GO
GO:0004725
GO:0016021
GO:0016787
GO:0050254
GO:0006470
GO:0008045
GO:0007399
GO:1903386
GO:0007412
GO:0048477
GO:0045467
GO:0048841
GO:0032093
GO:0007411
GO:0009925
GO:0120034
GO:0030424
GO:0008594
GO:0051491
GO:0007283
GO:0031290
GO:0060269
GO:0031252
GO:0008201
GO:0048675
GO:0035335
GO:0007155
GO:0051124
GO:0005001
GO:0005925
GO:0005515
GO:0007165
GO:0009452
GO:0004930
GO:0006355
GO:0043565
GO:0016567
GO:0016020
GO:0005634
GO:0006260
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 24,
Likelihood: 0.997773
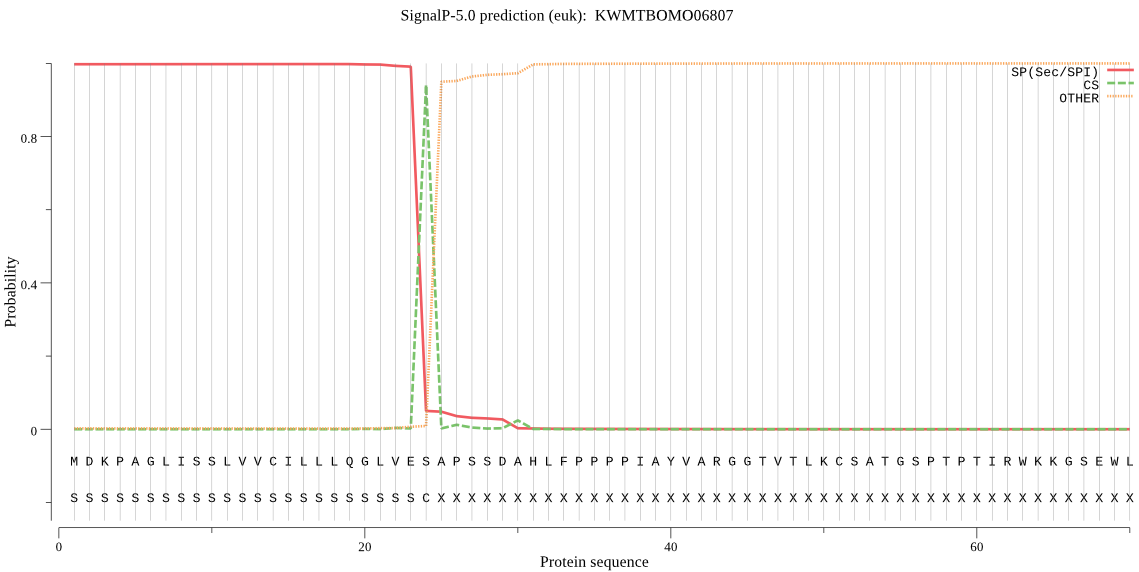
Length:
210
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.53240999999999
Exp number, first 60 AAs:
5.52757
Total prob of N-in:
0.28400
outside
1 - 210
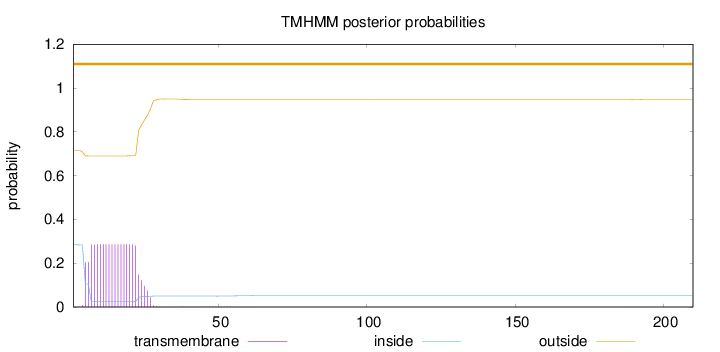
Population Genetic Test Statistics
Pi
13.458753
Theta
16.561996
Tajima's D
-0.945147
CLR
15.883075
CSRT
0.149842507874606
Interpretation
Uncertain
