Gene
KWMTBOMO06806
Pre Gene Modal
BGIBMGA012047
Annotation
PREDICTED:_protein_FAM195A-like_[Plutella_xylostella]
Full name
MAPK regulated corepressor interacting protein 2
Alternative Name
Protein FAM195A
Location in the cell
Nuclear Reliability : 1.861
Sequence
CDS
ATGAGCACTGGAATTATTTTTCACAGATCAAGTTATCATGATGATCTCATCAATTACATTTATGATTCGTGGAATAAGGTGACCCGTGATCTAGAACGTGGCGATGATGAGGCGAAGTACTACCATGACGTGATGACACCTCGTCACCTTGCCAACTTCAGGCCATTCAATCTTGATGAGTGGTGGGCGCGTCAGACCCACAATCGCAACAAGCATCGGTCTTAG
Protein
MSTGIIFHRSSYHDDLINYIYDSWNKVTRDLERGDDEAKYYHDVMTPRHLANFRPFNLDEWWARQTHNRNKHRS
Summary
Subunit
Interacts with DDX6. Interacts with MCRIP1.
Similarity
Belongs to the MCRIP family.
Keywords
Acetylation
Complete proteome
Cytoplasm
Methylation
Nucleus
Phosphoprotein
Reference proteome
Feature
chain MAPK regulated corepressor interacting protein 2
Uniprot
A0A194R4B0
A0A3S2LCF5
I4DR78
I4DQ82
A0A2A4JZ79
A0A2H1W218
+ More
A0A1E1WFE5 S4PW99 A0A212ESZ8 A0A0T6B0G0 V5GDK7 A0A158P191 E0VGS4 A0A154PSN4 A0A195B2V6 A0A1Q3F773 A0A1Q3F560 D6WGV5 E2AP35 F4WBH3 A0A195D8L5 A0A195FU92 A0A195DLI6 A0A151WWI9 A0A067QZ51 A0A023EFX4 A0A182GKR7 B0WRP2 A0A310SFF9 A0A182MZ65 V9IIL6 A0A088A9N7 Q0IEA1 E2BQS7 A0A3L8E346 A0A1B6MEF0 A0A026WEQ5 A0A2U9CQS2 A0A0M8ZW76 A0A1L8DFS3 A0A0A9YRV3 A0A182RDT4 A0A182Y0C3 A0A182MTM5 A0A182S8U3 A0A2P8ZMC7 A0A1B6ID87 A0A1B6FPP2 J9KVZ3 C4WSC1 A0A1A6HWS8 A0A182J3V4 A0A2H8TXN1 A0A3B4YKF6 A0A2S2QKG8 D7EHQ5 A0A067R3Z2 A0A3B4U9D0 A0A084W7E3 A0A182W6D8 A0A2S2PU28 E9H1M8 A0A182XNQ9 A0A1S4HE64 A0A1Y1M923 A0A182LMT6 K7FK69 M3W9C7 U5ER39 A0A0P5TCK6 A0A182PWB9 B4KHK3 A0A3Q3GPI0 A0A1S3JXV5 H2NPK7 Q9CQB2 G3PBP3 A0A1U7QY26 A0A224Y0A7 A0A0V0GB52 A0A023F8V7 A0A069DP94 F7D1U8 A0A1U8CMQ9 A0A0B6ZYE1 A0A195FH17 G3W7X7 A0A2J8R1H1 A0A0P4VN53 R4G4D2 A0A3Q1FEA6 A0A0B6ZZ11 I3M365 A0A3Q0DN40 A0A195DJ26 A0A1B6CSM9 A0A151I0T3 A0A151XHE8 A0A2L2Y3L4
A0A1E1WFE5 S4PW99 A0A212ESZ8 A0A0T6B0G0 V5GDK7 A0A158P191 E0VGS4 A0A154PSN4 A0A195B2V6 A0A1Q3F773 A0A1Q3F560 D6WGV5 E2AP35 F4WBH3 A0A195D8L5 A0A195FU92 A0A195DLI6 A0A151WWI9 A0A067QZ51 A0A023EFX4 A0A182GKR7 B0WRP2 A0A310SFF9 A0A182MZ65 V9IIL6 A0A088A9N7 Q0IEA1 E2BQS7 A0A3L8E346 A0A1B6MEF0 A0A026WEQ5 A0A2U9CQS2 A0A0M8ZW76 A0A1L8DFS3 A0A0A9YRV3 A0A182RDT4 A0A182Y0C3 A0A182MTM5 A0A182S8U3 A0A2P8ZMC7 A0A1B6ID87 A0A1B6FPP2 J9KVZ3 C4WSC1 A0A1A6HWS8 A0A182J3V4 A0A2H8TXN1 A0A3B4YKF6 A0A2S2QKG8 D7EHQ5 A0A067R3Z2 A0A3B4U9D0 A0A084W7E3 A0A182W6D8 A0A2S2PU28 E9H1M8 A0A182XNQ9 A0A1S4HE64 A0A1Y1M923 A0A182LMT6 K7FK69 M3W9C7 U5ER39 A0A0P5TCK6 A0A182PWB9 B4KHK3 A0A3Q3GPI0 A0A1S3JXV5 H2NPK7 Q9CQB2 G3PBP3 A0A1U7QY26 A0A224Y0A7 A0A0V0GB52 A0A023F8V7 A0A069DP94 F7D1U8 A0A1U8CMQ9 A0A0B6ZYE1 A0A195FH17 G3W7X7 A0A2J8R1H1 A0A0P4VN53 R4G4D2 A0A3Q1FEA6 A0A0B6ZZ11 I3M365 A0A3Q0DN40 A0A195DJ26 A0A1B6CSM9 A0A151I0T3 A0A151XHE8 A0A2L2Y3L4
Pubmed
26354079
22651552
23622113
22118469
21347285
20566863
+ More
18362917 19820115 20798317 21719571 24845553 24945155 26483478 17510324 30249741 24508170 25401762 26823975 25244985 29403074 24438588 21292972 12364791 28004739 20966253 17381049 17975172 17994087 16141072 15489334 17242355 19144319 21183079 24129315 25474469 26334808 17495919 21709235 27129103 26561354
18362917 19820115 20798317 21719571 24845553 24945155 26483478 17510324 30249741 24508170 25401762 26823975 25244985 29403074 24438588 21292972 12364791 28004739 20966253 17381049 17975172 17994087 16141072 15489334 17242355 19144319 21183079 24129315 25474469 26334808 17495919 21709235 27129103 26561354
EMBL
KQ460779
KPJ12359.1
RSAL01000045
RVE50610.1
AK404983
BAM20418.1
+ More
AK403977 KQ459252 BAM20072.1 KPJ02107.1 NWSH01000397 PCG76692.1 ODYU01005732 SOQ46872.1 GDQN01005398 GDQN01003634 JAT85656.1 JAT87420.1 GAIX01007158 JAA85402.1 AGBW02012669 OWR44618.1 LJIG01016369 KRT80809.1 GALX01006382 JAB62084.1 ADTU01006069 DS235151 EEB12580.1 KQ435152 KZC14912.1 KQ976641 KYM78813.1 GFDL01011631 JAV23414.1 GFDL01012335 JAV22710.1 KQ971321 EFA00594.1 GL441439 EFN64867.1 GL888063 EGI68452.1 KQ976749 KYN08769.1 KQ981272 KYN43862.1 KQ980765 KYN13359.1 KQ982691 KYQ52208.1 KK852815 KDR15824.1 GAPW01005873 JAC07725.1 JXUM01070453 KQ562607 KXJ75495.1 DS232058 EDS33446.1 KQ761906 OAD56451.1 JR047686 AEY60482.1 CH477899 EAT35235.1 GL449799 EFN81952.1 QOIP01000001 RLU26873.1 GEBQ01005718 JAT34259.1 KK107242 EZA54547.1 CP026261 AWP18931.1 KQ435841 KOX71306.1 GFDF01008874 JAV05210.1 GBHO01011344 GBRD01009795 GDHC01013976 JAG32260.1 JAG56029.1 JAQ04653.1 AXCM01001333 PYGN01000017 PSN57643.1 GECU01022827 JAS84879.1 GECZ01017592 JAS52177.1 ABLF02016982 ABLF02016983 ABLF02016984 ABLF02016986 AK340131 BAH70791.1 LZPO01008003 OBS82696.1 GFXV01006884 MBW18689.1 GGMS01009005 MBY78208.1 DS497665 EFA12137.1 KK852999 KDR12662.1 ATLV01021228 KE525315 KFB46137.1 GGMR01020268 MBY32887.1 GL732583 EFX74472.1 AAAB01008984 GEZM01042484 JAV79827.1 AGCU01024909 AGCU01024910 AANG04000434 GANO01003918 JAB55953.1 GDIQ01219573 GDIP01128012 LRGB01000024 JAK32152.1 JAL75702.1 KZS21553.1 CH933807 EDW12282.1 ABGA01033509 ABGA01033510 ABGA01033511 AK003564 AK020614 BC024332 GFTR01002423 JAW14003.1 GECL01001156 JAP04968.1 GBBI01000841 JAC17871.1 GBGD01003383 JAC85506.1 HACG01026713 CEK73578.1 KQ981560 KYN39980.1 AEFK01080146 AEFK01080147 NDHI03003828 PNJ02356.1 GDKW01000551 JAI56044.1 ACPB03028992 GAHY01000871 JAA76639.1 HACG01026712 CEK73577.1 AGTP01030746 KQ980800 KYN12822.1 GEDC01020897 GEDC01018294 JAS16401.1 JAS19004.1 KQ976599 KYM79502.1 KQ982130 KYQ59826.1 IAAA01005613 LAA02741.1
AK403977 KQ459252 BAM20072.1 KPJ02107.1 NWSH01000397 PCG76692.1 ODYU01005732 SOQ46872.1 GDQN01005398 GDQN01003634 JAT85656.1 JAT87420.1 GAIX01007158 JAA85402.1 AGBW02012669 OWR44618.1 LJIG01016369 KRT80809.1 GALX01006382 JAB62084.1 ADTU01006069 DS235151 EEB12580.1 KQ435152 KZC14912.1 KQ976641 KYM78813.1 GFDL01011631 JAV23414.1 GFDL01012335 JAV22710.1 KQ971321 EFA00594.1 GL441439 EFN64867.1 GL888063 EGI68452.1 KQ976749 KYN08769.1 KQ981272 KYN43862.1 KQ980765 KYN13359.1 KQ982691 KYQ52208.1 KK852815 KDR15824.1 GAPW01005873 JAC07725.1 JXUM01070453 KQ562607 KXJ75495.1 DS232058 EDS33446.1 KQ761906 OAD56451.1 JR047686 AEY60482.1 CH477899 EAT35235.1 GL449799 EFN81952.1 QOIP01000001 RLU26873.1 GEBQ01005718 JAT34259.1 KK107242 EZA54547.1 CP026261 AWP18931.1 KQ435841 KOX71306.1 GFDF01008874 JAV05210.1 GBHO01011344 GBRD01009795 GDHC01013976 JAG32260.1 JAG56029.1 JAQ04653.1 AXCM01001333 PYGN01000017 PSN57643.1 GECU01022827 JAS84879.1 GECZ01017592 JAS52177.1 ABLF02016982 ABLF02016983 ABLF02016984 ABLF02016986 AK340131 BAH70791.1 LZPO01008003 OBS82696.1 GFXV01006884 MBW18689.1 GGMS01009005 MBY78208.1 DS497665 EFA12137.1 KK852999 KDR12662.1 ATLV01021228 KE525315 KFB46137.1 GGMR01020268 MBY32887.1 GL732583 EFX74472.1 AAAB01008984 GEZM01042484 JAV79827.1 AGCU01024909 AGCU01024910 AANG04000434 GANO01003918 JAB55953.1 GDIQ01219573 GDIP01128012 LRGB01000024 JAK32152.1 JAL75702.1 KZS21553.1 CH933807 EDW12282.1 ABGA01033509 ABGA01033510 ABGA01033511 AK003564 AK020614 BC024332 GFTR01002423 JAW14003.1 GECL01001156 JAP04968.1 GBBI01000841 JAC17871.1 GBGD01003383 JAC85506.1 HACG01026713 CEK73578.1 KQ981560 KYN39980.1 AEFK01080146 AEFK01080147 NDHI03003828 PNJ02356.1 GDKW01000551 JAI56044.1 ACPB03028992 GAHY01000871 JAA76639.1 HACG01026712 CEK73577.1 AGTP01030746 KQ980800 KYN12822.1 GEDC01020897 GEDC01018294 JAS16401.1 JAS19004.1 KQ976599 KYM79502.1 KQ982130 KYQ59826.1 IAAA01005613 LAA02741.1
Proteomes
UP000053240
UP000283053
UP000053268
UP000218220
UP000007151
UP000005205
+ More
UP000009046 UP000076502 UP000078540 UP000007266 UP000000311 UP000007755 UP000078542 UP000078541 UP000078492 UP000075809 UP000027135 UP000069940 UP000249989 UP000002320 UP000075884 UP000005203 UP000008820 UP000008237 UP000279307 UP000053097 UP000246464 UP000053105 UP000075900 UP000076408 UP000075883 UP000075901 UP000245037 UP000007819 UP000092124 UP000075880 UP000261360 UP000261420 UP000030765 UP000075920 UP000000305 UP000076407 UP000075882 UP000007267 UP000011712 UP000076858 UP000075885 UP000009192 UP000261660 UP000085678 UP000001595 UP000000589 UP000007635 UP000189706 UP000002280 UP000007648 UP000015103 UP000257200 UP000005215 UP000189704
UP000009046 UP000076502 UP000078540 UP000007266 UP000000311 UP000007755 UP000078542 UP000078541 UP000078492 UP000075809 UP000027135 UP000069940 UP000249989 UP000002320 UP000075884 UP000005203 UP000008820 UP000008237 UP000279307 UP000053097 UP000246464 UP000053105 UP000075900 UP000076408 UP000075883 UP000075901 UP000245037 UP000007819 UP000092124 UP000075880 UP000261360 UP000261420 UP000030765 UP000075920 UP000000305 UP000076407 UP000075882 UP000007267 UP000011712 UP000076858 UP000075885 UP000009192 UP000261660 UP000085678 UP000001595 UP000000589 UP000007635 UP000189706 UP000002280 UP000007648 UP000015103 UP000257200 UP000005215 UP000189704
PRIDE
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A194R4B0
A0A3S2LCF5
I4DR78
I4DQ82
A0A2A4JZ79
A0A2H1W218
+ More
A0A1E1WFE5 S4PW99 A0A212ESZ8 A0A0T6B0G0 V5GDK7 A0A158P191 E0VGS4 A0A154PSN4 A0A195B2V6 A0A1Q3F773 A0A1Q3F560 D6WGV5 E2AP35 F4WBH3 A0A195D8L5 A0A195FU92 A0A195DLI6 A0A151WWI9 A0A067QZ51 A0A023EFX4 A0A182GKR7 B0WRP2 A0A310SFF9 A0A182MZ65 V9IIL6 A0A088A9N7 Q0IEA1 E2BQS7 A0A3L8E346 A0A1B6MEF0 A0A026WEQ5 A0A2U9CQS2 A0A0M8ZW76 A0A1L8DFS3 A0A0A9YRV3 A0A182RDT4 A0A182Y0C3 A0A182MTM5 A0A182S8U3 A0A2P8ZMC7 A0A1B6ID87 A0A1B6FPP2 J9KVZ3 C4WSC1 A0A1A6HWS8 A0A182J3V4 A0A2H8TXN1 A0A3B4YKF6 A0A2S2QKG8 D7EHQ5 A0A067R3Z2 A0A3B4U9D0 A0A084W7E3 A0A182W6D8 A0A2S2PU28 E9H1M8 A0A182XNQ9 A0A1S4HE64 A0A1Y1M923 A0A182LMT6 K7FK69 M3W9C7 U5ER39 A0A0P5TCK6 A0A182PWB9 B4KHK3 A0A3Q3GPI0 A0A1S3JXV5 H2NPK7 Q9CQB2 G3PBP3 A0A1U7QY26 A0A224Y0A7 A0A0V0GB52 A0A023F8V7 A0A069DP94 F7D1U8 A0A1U8CMQ9 A0A0B6ZYE1 A0A195FH17 G3W7X7 A0A2J8R1H1 A0A0P4VN53 R4G4D2 A0A3Q1FEA6 A0A0B6ZZ11 I3M365 A0A3Q0DN40 A0A195DJ26 A0A1B6CSM9 A0A151I0T3 A0A151XHE8 A0A2L2Y3L4
A0A1E1WFE5 S4PW99 A0A212ESZ8 A0A0T6B0G0 V5GDK7 A0A158P191 E0VGS4 A0A154PSN4 A0A195B2V6 A0A1Q3F773 A0A1Q3F560 D6WGV5 E2AP35 F4WBH3 A0A195D8L5 A0A195FU92 A0A195DLI6 A0A151WWI9 A0A067QZ51 A0A023EFX4 A0A182GKR7 B0WRP2 A0A310SFF9 A0A182MZ65 V9IIL6 A0A088A9N7 Q0IEA1 E2BQS7 A0A3L8E346 A0A1B6MEF0 A0A026WEQ5 A0A2U9CQS2 A0A0M8ZW76 A0A1L8DFS3 A0A0A9YRV3 A0A182RDT4 A0A182Y0C3 A0A182MTM5 A0A182S8U3 A0A2P8ZMC7 A0A1B6ID87 A0A1B6FPP2 J9KVZ3 C4WSC1 A0A1A6HWS8 A0A182J3V4 A0A2H8TXN1 A0A3B4YKF6 A0A2S2QKG8 D7EHQ5 A0A067R3Z2 A0A3B4U9D0 A0A084W7E3 A0A182W6D8 A0A2S2PU28 E9H1M8 A0A182XNQ9 A0A1S4HE64 A0A1Y1M923 A0A182LMT6 K7FK69 M3W9C7 U5ER39 A0A0P5TCK6 A0A182PWB9 B4KHK3 A0A3Q3GPI0 A0A1S3JXV5 H2NPK7 Q9CQB2 G3PBP3 A0A1U7QY26 A0A224Y0A7 A0A0V0GB52 A0A023F8V7 A0A069DP94 F7D1U8 A0A1U8CMQ9 A0A0B6ZYE1 A0A195FH17 G3W7X7 A0A2J8R1H1 A0A0P4VN53 R4G4D2 A0A3Q1FEA6 A0A0B6ZZ11 I3M365 A0A3Q0DN40 A0A195DJ26 A0A1B6CSM9 A0A151I0T3 A0A151XHE8 A0A2L2Y3L4
Ontologies
Topology
Subcellular location
Cytoplasm
Upon cellular stress, relocalizes to stress granules. With evidence from 3 publications.
Stress granule Upon cellular stress, relocalizes to stress granules. With evidence from 3 publications.
Nucleus Upon cellular stress, relocalizes to stress granules. With evidence from 3 publications.
Stress granule Upon cellular stress, relocalizes to stress granules. With evidence from 3 publications.
Nucleus Upon cellular stress, relocalizes to stress granules. With evidence from 3 publications.
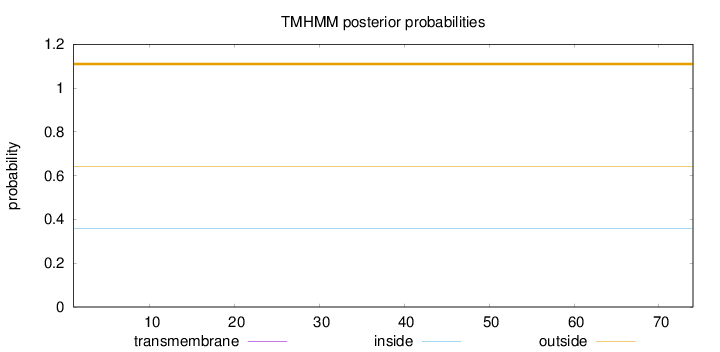
Length:
74
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.35898
outside
1 - 74
Population Genetic Test Statistics
Pi
25.7402
Theta
22.160983
Tajima's D
0.59356
CLR
0.943891
CSRT
0.546972651367432
Interpretation
Uncertain
